Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila
- PMID: 9637685
- PMCID: PMC316910
- DOI: 10.1101/gad.12.12.1837
Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila
Abstract
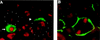
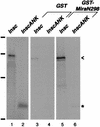
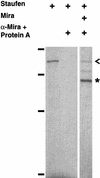
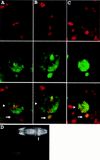
Neuroblasts in the developing Drosophila CNS asymmetrically localize the cell fate determinants Numb and Prospero as well as prospero RNA to the basal cortex during mitosis. The localization of Prospero requires the function of inscuteable and miranda, whereas prospero RNA localization requires inscuteable and staufen function. We demonstrate that Miranda contains multiple functional domains: an amino-terminal asymmetric localization domain, which interacts with Inscuteable, a central Numb interaction domain, and a more carboxy-terminal Prospero interaction domain. We also show that Miranda and Staufen have similar subcellular localization patterns and interact in vitro. Furthermore, miranda function is required for the asymmetric localization of Staufen. Miranda localization is disrupted by the microfilament disrupting agent latrunculin A. Our results suggest that Miranda directs the basal cortical localization of multiple molecules, including Staufen and prospero RNA, in mitotic neuroblasts in an actin-dependent manner.
Figures




Similar articles
-
Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release.Mol Cell Neurosci. 1998 Dec;12(6):325-39. doi: 10.1006/mcne.1998.0724. Mol Cell Neurosci. 1998. PMID: 9888987
-
Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts.Curr Biol. 1997 Nov 1;7(11):827-35. doi: 10.1016/s0960-9822(06)00370-8. Curr Biol. 1997. PMID: 9382803
-
Asymmetry and cell fate in the Drosophila embryonic CNS.Int J Dev Biol. 1998;42(3):379-83. Int J Dev Biol. 1998. PMID: 9654022 Review.
-
Miranda mediates asymmetric protein and RNA localization in the developing nervous system.Genes Dev. 1998 Jun 15;12(12):1847-57. doi: 10.1101/gad.12.12.1847. Genes Dev. 1998. PMID: 9637686 Free PMC article.
-
Asymmetic division: dynastic intricacies of neuroblast division.Curr Biol. 1997 Nov 1;7(11):R726-8. doi: 10.1016/s0960-9822(06)00367-8. Curr Biol. 1997. PMID: 9382791 Review.
Cited by
-
Drosophila as a Model for Developmental Biology: Stem Cell-Fate Decisions in the Developing Nervous System.J Dev Biol. 2018 Oct 19;6(4):25. doi: 10.3390/jdb6040025. J Dev Biol. 2018. PMID: 30347666 Free PMC article. Review.
-
Stem cell regulation. Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency.Science. 2015 Nov 20;350(6263):aab0988. doi: 10.1126/science.aab0988. Science. 2015. PMID: 26586765 Free PMC article.
-
Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts.J Cell Biol. 2000 Sep 18;150(6):1361-74. doi: 10.1083/jcb.150.6.1361. J Cell Biol. 2000. PMID: 10995441 Free PMC article.
-
aPKC-mediated displacement and actomyosin-mediated retention polarize Miranda in Drosophila neuroblasts.Elife. 2018 Jan 24;7:e29939. doi: 10.7554/eLife.29939. Elife. 2018. PMID: 29364113 Free PMC article.
-
Insights into mRNA transport in neurons.Proc Natl Acad Sci U S A. 2003 Feb 18;100(4):1465-6. doi: 10.1073/pnas.0630376100. Epub 2003 Feb 10. Proc Natl Acad Sci U S A. 2003. PMID: 12578967 Free PMC article. No abstract available.
References
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Broadus J, Doe CQ. Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr Biol. 1997;7:827–835. - PubMed
-
- Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;39:792–795. - PubMed
-
- Burchard S, Paululat A, Hinz U, Renkawitz-Pohl R. The mutant not enough muscles (nem) reveals reduction of the Drosophila embryonic muscle pattern. J Cell Sci. 1995;108:1443–1454. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
