Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine
- PMID: 9642198
- PMCID: PMC107300
- DOI: 10.1128/JB.180.13.3432-3440.1998
Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine
Abstract
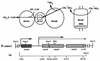
Coenzyme M (CoM) is methylated during methanogenesis from monomethyamine in a reaction catalyzed by three proteins. Using monomethylamine, a 52-kDa polypeptide termed monomethylamine methyltransferase (MMAMT) methylates the corrinoid cofactor bound to a second polypeptide, monomethylamine corrinoid protein (MMCP). Methylated MMCP then serves as a substrate for MT2-A, which methylates CoM. The genes for these proteins are clustered on 6.8 kb of DNA in Methanosarcina barkeri MS. The gene encoding MMCP (mtmC) is located directly upstream of the gene encoding MMAMT (mtmB). The gene encoding MT2-A (mtbA) was found 1.1 kb upstream of mtmC, but no obvious open reading frame was found in the intergenic region between mtbA and mtmC. A single monocistronic transcript was found for mtbA that initiated 76 bp from the translational start. Separate transcripts of 2.4 and 4.7 kb were detected, both of which carried mtmCB. The larger transcript also encoded mtmP, which is homologous to the APC family of cationic amine permeases and may therefore encode a methylamine permease. A single transcriptional start site was found 447 bp upstream of the translational start of mtmC. MtmC possesses the corrinoid binding motif found in corrinoid proteins involved in dimethylsulfide- and methanol-dependent methanogenesis, as well as in methionine synthase. The open reading frame of mtmB was interrupted by a single in-frame, midframe, UAG codon which was also found in mtmB from M. barkeri NIH. A mechanism that circumvents UAG-directed termination of translation must operate during expression of mtmB in this methanogen.
Figures






References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Boone D R, Whitman W B, Rouvière P. Methanogenesis. Ecology, physiology, biochemistry, and genetics. New York, N.Y: Chapman & Hall; 1993. Diversity and taxonomy of methanogens; pp. 35–80.
-
- Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodeck A, Scott J L, Georghagen N S M, Weidman J F, Fuhrman J L, Nguyen D, Utterback T R, Kelly J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C F, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
-
- Burgett S G, Rosteck P R. Use of dimethylsulfoxide in fluorescent tag sequencing. In: Adams M D, Fields C, Venter J C, editors. Automated DNA sequencing and analysis. San Diego, Calif: Academic Press; 1994. pp. 211–215.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

