Recognition of a human arrest site is conserved between RNA polymerase II and prokaryotic RNA polymerases
- PMID: 9642244
- PMCID: PMC3371603
- DOI: 10.1074/jbc.273.27.16843
Recognition of a human arrest site is conserved between RNA polymerase II and prokaryotic RNA polymerases
Abstract
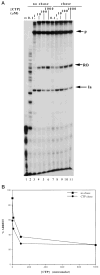
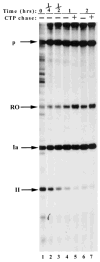
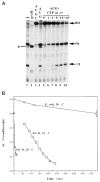
DNA sequences that arrest transcription by either eukaryotic RNA polymerase II or Escherichia coli RNA polymerase have been identified previously. Elongation factors SII and GreB are RNA polymerase-binding proteins that enable readthrough of arrest sites by these enzymes, respectively. This functional similarity has led to general models of elongation applicable to both eukaryotic and prokaryotic enzymes. Here we have transcribed with phage and bacterial RNA polymerases, a human DNA sequence previously defined as an arrest site for RNA polymerase II. The phage and bacterial enzymes both respond efficiently to the arrest signal in vitro at limiting levels of nucleoside triphosphates. The E. coli polymerase remains in a template-engaged complex for many hours, can be isolated, and is potentially active. The enzyme displays a relatively slow first-order loss of elongation competence as it dwells at the arrest site. Bacterial RNA polymerase arrested at the human site is reactivated by GreB in the same way that RNA polymerase II arrested at this site is stimulated by SII. Very efficient readthrough can be achieved by phage, bacterial, and eukaryotic RNA polymerases in the absence of elongation factors if 5-Br-UTP is substituted for UTP. These findings provide additional and direct evidence for functional similarity between prokaryotic and eukaryotic transcription elongation and readthrough mechanisms.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

