A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans
- PMID: 9647650
- PMCID: PMC2133011
- DOI: 10.1083/jcb.141.7.1575
A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans
Abstract
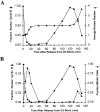
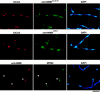
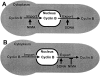
NIMA promotes entry into mitosis in late G2 by some mechanism that is after activation of the Aspergillus nidulans G2 cyclin-dependent kinase, NIMXCDC2/NIMECyclin B. Here we present two independent lines of evidence which indicate that this mechanism involves control of NIMXCDC2/NIMECyclin B localization. First, we found that NIMECyclin B localized to the nucleus and the nucleus-associated organelle, the spindle pole body, in a NIMA-dependent manner. Analysis of cells from asynchronous cultures, synchronous cultures, and cultures arrested in S or G2 showed that NIMECyclin B was predominantly nuclear during interphase, with maximal nuclear accumulation in late G2. NIMXCDC2 colocalized with NIMECyclin B in G2 cells. Although inactivation of NIMA using either the nimA1 or nimA5 temperature-sensitive mutations blocked cells in G2, NIMXCDC2/NIMECyclin B localization was predominantly cytoplasmic rather than nuclear. Second, we found that nimA interacts genetically with sonA, which is a homologue of the yeast nucleocytoplasmic transporter GLE2/RAE1. Mutations in sonA were identified as allele-specific suppressors of nimA1. The sonA1 suppressor alleviated the nuclear division and NIMECyclin B localization defects of nimA1 cells without markedly increasing NIMXCDC2 or NIMA kinase activity. These results indicate that NIMA promotes the nuclear localization of the NIMXCDC2/ NIMECyclin B complex, by a process involving SONA. This mechanism may be involved in coordinating the functions of NIMXCDC2 and NIMA in the regulation of mitosis.
Figures










References
-
- Alfa CE, Ducommun B, Beach D, Hyams JS. Distinct nuclear and spindle pole body populations of cyclincdc2in fission yeast. Nature. 1990;347:680–682. - PubMed
-
- Alfa CE, Gallagher IM, Hyams JS. Subcellular localization of the p34cdc2/p63cdc13protein kinase in fission yeast. Cold Spring Harbor Symp Quant Biol. 1991;56:489–494. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Audit M, Barbier M, Soyer-Gobillard MO, Albert M, Geraud ML, Nicolas G, Lenaers G. Cyclin B (p56cdc13) localization in the yeast Schizosaccharomyces pombe: an ultrastructural and immunocytochemical study. Biol Cell. 1996;86:1–10. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

