New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2
- PMID: 9649510
- PMCID: PMC316977
- DOI: 10.1101/gad.12.13.2073
New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2
Abstract
The POU transcription factor Oct-4 is expressed specifically in the germ line, pluripotent cells of the pregastrulation embryo and stem cell lines derived from the early embryo. Osteopontin (OPN) is a protein secreted by cells of the preimplantation embryo and contains a GRGDS motif that can bind to specific integrin subtypes and modulate cell adhesion/migration. We show that Oct-4 and OPN are coexpressed in the preimplantation mouse embryo and during differentiation of embryonal cell lines. Immunoprecipitation of the first intron of OPN (i-opn) from covalently fixed chromatin of embryonal stem cells by Oct-4-specific antibodies indicates that Oct-4 binds to this fragment in vivo. The i-opn fragment functions as an enhancer in cell lines that resemble cells of the preimplantation embryo. Furthermore, it contains a novel palindromic Oct factor recognition element (PORE) that is composed of an inverted pair of homeodomain-binding sites separated by exactly 5 bp (ATTTG +5 CAAAT). POU proteins can homo- and heterodimerize on the PORE in a configuration that has not been described previously. Strong transcriptional activation of the OPN element requires an intact PORE. In contrast, the canonical octamer overlapping with the downstream half of the PORE is not essential. Sox-2 is a transcription factor that contains an HMG box and is coexpressed with Oct-4 in the early mouse embryo. Sox-2 represses Oct-4 mediated activation of i-opn by way of a canonical Sox element that is located close to the PORE. Repression depends on a carboxy-terminal region of Sox-2 that is outside of the HMG box. Expression, DNA binding, and transactivation data are consistent with the hypothesis that OPN expression is regulated by Oct-4 and Sox-2 in preimplantation development.
Figures

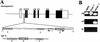


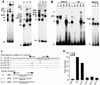
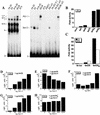
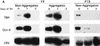


References
-
- Adamson ED. Cell-lineage-specific gene expression in development. In: Rossant J, Pederson RA, editors. Experimental approaches to mammalian embryonic development. Cambridge, UK: Cambridge University Press; 1986. pp. 321–364.
-
- Behrendtsen O, Alexander CM, Werb Z. Cooperative interactions between extracellular matrix, integrins and parathyroid hormone-related peptide regulate parietal endoderm differentiation in mouse embryos. Development. 1995;121:4137–4148. - PubMed
-
- Chinea G, Padron G, Hooft RW, Sander C, Vriend G. The use of position-specific rotamers in model building by homology. Proteins. 1995;23:415–421. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
