Utility of random amplified polymorphic DNA PCR and TaqMan automated detection in molecular identification of Aspergillus fumigatus
- PMID: 9650962
- PMCID: PMC104978
- DOI: 10.1128/JCM.36.7.2057-2062.1998
Utility of random amplified polymorphic DNA PCR and TaqMan automated detection in molecular identification of Aspergillus fumigatus
Abstract
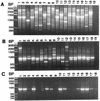
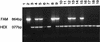
We developed a method for the identification of Aspergillus fumigatus fungal isolates by using random amplified polymorphic DNA (RAPD) PCR (RAPD-PCR) cloning and the TaqMan LS50B fluorogenic detection system (Perkin-Elmer Corp., Applied Biosystems, Foster City, Calif.). DNA from seven clinically important Aspergillus species was screened by RAPD-PCR to identify section- or species-specific amplicons. With the OPZ19 RAPD primer a 1,264-bp product was amplified from all A. fumigatus strains initially examined but not from other species. A partial DNA sequence of this product was used to design a specific primer pair, which generated a single 864-bp fragment with DNA from 90 of 100 A. fumigatus isolates when a "touchdown" (65-->55 degrees C) annealing protocol was used. The TaqMan system, a fluorogenic assay which uses the 5'-->3' endonuclease activity of Taq DNA polymerase, detected this 864-bp product with DNA from 89 of these 90 A. fumigatus strains; 1 DNA sample generated an indeterminate result. With DNA from three morphologically typical A. fumigatus isolates, six white ("albino") A. fumigatus isolates, and five of six Neosartorya species (non-A. fumigatus members of the section Fumigati), the 864-bp product was amplified differentially at an annealing temperature of 56 degrees C but not with the touchdown annealing format. No amplicon was detected with DNA from 56 isolates of heterologous Aspergillus, Penicillium, and Paecilomyces species or from Neosartorya fennelliae; TaqMan assay results were either negative (51 isolates) or indeterminate (5 isolates) for all isolates. This RAPD-PCR and TaqMan assay offers promise as a nucleic acid-based system that can be used for the identification of filamentous fungal isolates and that requires no postamplification sample manipulations.
Figures



References
-
- Andriole, V. T. 1993. Infections with Aspergillus species. Clin. Infect. Dis. 17(Suppl. 2):S481–S486. - PubMed
-
- Armitage P, Berry G. Statistical methods in medical research. 2nd ed. London, United Kingdom: Blackwell Scientific Publications; 1987.
-
- Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991.
-
- Brandt M E, Hutwagner L C, Kuykendall R J, Pinner R W the Cryptococcal Disease Active Surveillance Group. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. J Clin Microbiol. 1995;33:1890–1895. - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

