The gut-enriched Krüppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion
- PMID: 9651398
- PMCID: PMC2275057
- DOI: 10.1074/jbc.273.28.17917
The gut-enriched Krüppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion
Abstract
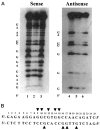
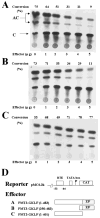
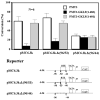
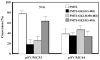
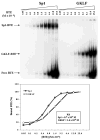
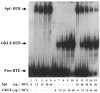
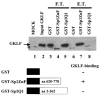
The gut-enriched Krüppel-like factor (GKLF) is a newly identified zinc finger-containing transcription factor. Recent studies indicate that GKLF binds to a core DNA sequence of 5'-(G/A)(G/A)GG(C/T)G(C/T)-3', which is found in an endogenous cis element, the basic transcription element (BTE) of the cytochrome P-450IA1 (CYP1A1) promoter. The present study characterizes the ability of GKLF to regulate CYP1A1 expression. By electrophoretic mobility gel shift assay (EMSA) and methylation interference assay, GKLF was found to bind BTE in a manner similar to several other transcription factors known to interact with BTE including Sp1 and BTEB. Cotransfection studies in Chinese hamster ovary cells showed that GKLF inhibited the CYP1A1 promoter in a dose- and BTE-dependent manner. The same experiments also revealed that BTE was responsible for a significant portion of the CYP1A1 promoter activity. EMSA of nuclear extracts from Chinese hamster ovary cells showed that Sp1 and Sp3 were two major proteins that interacted with BTE. Additional cotransfection studies showed that GKLF inhibited Sp1-mediated activation of the CYP1A1 promoter. In contrast, GKLF enhanced Sp3-dependent suppression of the same promoter. Moreover, the ability of GKLF to inhibit Sp1-dependent transactivation was in part due to physical interaction of the two proteins. These findings indicate that GKLF is a negative regulator of the CYP1A1 promoter in a BTE-dependent fashion and that this inhibitory effect is in part mediated by physical interaction with Sp1.
Figures





References
-
- Klug A, Rhodes D. Cold Spring Harb Symp Quant Biol. 1987;52:473–482. - PubMed
-
- Berg JM. J Biol Chem. 1990;265:6513–6516. - PubMed
-
- Berg JM. Annu Rev Biophys Biophys Chem. 1990;19:405–421. - PubMed
-
- Klug A, Schwabe JW. FASEB J. 1995;9:597–604. - PubMed
-
- Berg JM, Shi Y. Science. 1996;271:1081–1085. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

