Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection
- PMID: 9653085
- PMCID: PMC2525546
- DOI: 10.1084/jem.188.1.71
Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection
Abstract
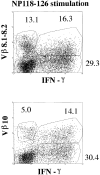
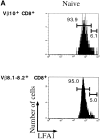
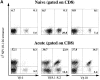
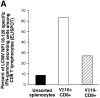
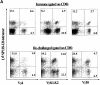
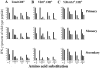
Viral infections often induce potent CD8 T cell responses that play a key role in antiviral immunity. After viral clearance, the vast majority of the expanded CD8 T cells undergo apoptosis, leaving behind a stable number of memory cells. The relationship between the CD8 T cells that clear the acute viral infection and the long-lived CD8 memory pool remaining in the individual is not fully understood. To address this issue, we examined the T cell receptor (TCR) repertoire of virus-specific CD8 T cells in the mouse model of infection with lymphocytic choriomeningitis virus (LCMV) using three approaches: (a) in vivo quantitative TCR beta chain V segment and complementarity determining region 3 (CDR3) length repertoire analysis by spectratyping (immunoscope); (b) identification of LCMV-specific CD8 T cells with MHC class I tetramers containing viral peptide and costaining with TCR Vbeta-specific antibodies; and (c) functional TCR fingerprinting based on recognition of variant peptides. We compared the repertoire of CD8 T cells responding to acute primary and secondary LCMV infections, together with that of virus-specific memory T cells in immune mice. Our analysis showed that CD8 T cells from several Vbeta families participated in the anti-LCMV response directed to the dominant cytotoxic T lymphocyte (CTL) epitope (NP118-126). However, the bulk (approximately 70%) of this CTL response was due to three privileged T cell populations systematically expanding during LCMV infection. Approximately 30% of the response consisted of Vbeta10+ CD8 T cells with a beta chain CDR3 length of nine amino acids, and 40% consisted of Vbeta8.1+ (beta CDR3 = eight amino acids) and Vbeta8.2+ cells (beta CDR3 = six amino acids). Finally, we showed that the TCR repertoire of the primary antiviral CD8 T cell response was similar both structurally and functionally to that of the memory pool and the secondary CD8 T cell effectors. These results suggest a stochastic selection of memory cells from the pool of CD8 T cells activated during primary infection.
Figures












References
-
- Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. - PubMed
-
- Wilson RK, Lai E, Concannon P, Barth RK, Hood LE. Structure, organization and polymorphism of murine and human T-cell receptor alpha and beta chain gene families. Immunol Rev. 1988;101:149–172. - PubMed
-
- Davis MM. T cell receptor gene diversity and selection. Annu Rev Biochem. 1990;59:475–496. - PubMed
-
- Goverman J, Minard K, Shastri N, Hunkapiller T, Hansburg D, Sercarz E, Hood L. Rearranged beta T cell receptor genes in a helper T cell clone specific for lysozyme: no correlation between V beta and MHC restriction. Cell. 1985;40:859–867. - PubMed
-
- Goverman J, Hunkapiller T, Hood L. A speculative view of the multicomponent nature of T cell antigen recognition. Cell. 1986;45:475–484. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

