The Notch1 receptor is cleaved constitutively by a furin-like convertase
- PMID: 9653148
- PMCID: PMC20937
- DOI: 10.1073/pnas.95.14.8108
The Notch1 receptor is cleaved constitutively by a furin-like convertase
Abstract
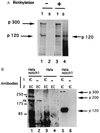
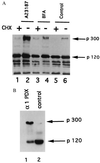
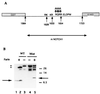
The Notch receptor, which is involved in numerous cell fate decisions in invertebrates and vertebrates, is synthesized as a 300-kDa precursor molecule (p300). We show here that proteolytic processing of p300 is an essential step in the formation of the biologically active receptor because only the cleaved fragments are present at the cell surface. Our results confirm and extend recent reports indicating that the Notch receptor exists at the plasma membrane as a heterodimeric molecule, but disagree as to the nature of the protease that is responsible for the cleavage that takes place in the extracellular region. We report here that constitutive processing of murine Notch1 involves a furin-like convertase. We show that the calcium ionophore A23187 and the alpha1-antitrypsin variant, alpha 1-PDX, a known inhibitor of furin-like convertases, inhibit p300 processing. When expressed in the furin-deficient Lovo cell line, p300 is not processed. In vitro digestion of a recombinant Notch-derived substrate with purified furin allowed mapping of the processing site to the carboxyl side of the sequence RQRR (amino acids 1651-1654). Mutation of these four amino acids (and of two secondary dibasic furin sites located nearby) completely abolished processing of the Notch1 receptor.
Figures
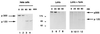

References
-
- Artavanis-Tsakonas S, Matsuno K, Fortini M E. Science. 1995;268:225–232. - PubMed
-
- Fleming R J, Purcell K, Artavanis-Tsakonas S. Trends Cell Biol. 1997;7:437–441. - PubMed
-
- Jarriault S, Brou C, Logeat F, Schroeter E, Kopan R, Israel A. Nature (London) 1995;377:355–358. - PubMed
-
- Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Genes Dev. 1993;7:1949–1965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

