Rice (Oryza sativa) centromeric regions consist of complex DNA
- PMID: 9653153
- PMCID: PMC20942
- DOI: 10.1073/pnas.95.14.8135
Rice (Oryza sativa) centromeric regions consist of complex DNA
Abstract
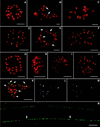
Rice bacterial artificial chromosome clones containing centromeric DNA were isolated by using a DNA sequence (pSau3A9) that is present in the centromeres of Gramineae species. Seven distinct repetitive DNA elements were isolated from a 75-kilobase rice bacterial artificial chromosome clone. All seven DNA elements are present in every rice centromere as demonstrated by fluorescence in situ hybridization. Six of the elements are middle repetitive, and their copy numbers range from approximately 50 to approximately 300 in the rice genome. Five of these six middle repetitive DNA elements are present in all of the Gramineae species, and the other element is detected only in species within the Bambusoideae subfamily of Gramineae. All six middle repetitive DNA elements are dispersed in the centromeric regions. The seventh element, the RCS2 family, is a tandem repeat of a 168-bp sequence that is represented approximately 6,000 times in the rice genome and is detected only in Oryza species. Fiber-fluorescence in situ hybridization analysis revealed that the RCS2 family is organized into long uninterrupted arrays and resembles previously reported tandem repeats located in the centromeres of human and Arabidopsis thaliana chromosomes. We characterized a large DNA fragment derived from a plant centromere and demonstrated that rice centromeres consist of complex DNA, including both highly and middle repetitive DNA sequences.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

