Normal retina releases a diffusible factor stimulating cone survival in the retinal degeneration mouse
- PMID: 9653191
- PMCID: PMC20980
- DOI: 10.1073/pnas.95.14.8357
Normal retina releases a diffusible factor stimulating cone survival in the retinal degeneration mouse
Abstract
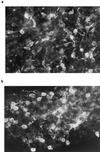
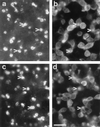
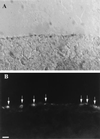
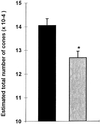
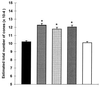
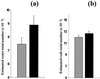
The role of cellular interactions in the mechanism of secondary cone photoreceptor degeneration in inherited retinal degenerations in which the mutation specifically affects rod photoreceptors was studied. We developed an organ culture model of whole retinas from 5-week-old mice carrying the retinal degeneration mutation, which at this age contain few remaining rods and numerous surviving cones cocultured with primary cultures of mixed cells from postnatal day 8 normal-sighted mice (C57BL/6) retinas or retinal explants from normal (C57BL/6) or dystrophic (C3H/He) 5-week-old mice. After 7 days, the numbers of residual cone photoreceptors were quantified after specific peanut lectin or anti-arrestin antibody labeling by using an unbiased stereological approach. Examination of organ cultured retinas revealed significantly greater numbers of surviving cones (15-20%) if cultured in the presence of retinas containing normal rods as compared with controls or cocultures with rod-deprived retinas. These data indicate the existence of a diffusible trophic factor released from retinas containing rod cells and acting on retinas in which only cones are present. Because cones are responsible for high acuity and color vision, such data could have important implications not only for eventual therapeutic approaches to human retinal degenerations but also to define interactions between retinal photoreceptor types.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

