Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene
- PMID: 9657992
- PMCID: PMC107317
- DOI: 10.1128/JB.180.14.3522-3528.1998
Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene
Abstract
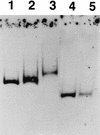
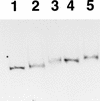
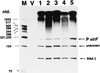
virF is the master regulator which activates the virulence determinant genes of Shigella spp. such as ipaBCD and virG. We previously reported that expression of virF itself is regulated in a pH-dependent manner and that cpxA, a sensor of a two-component regulatory system, is involved in this regulation (S. Nakayama and H. Watanabe, J. Bacteriol. 177:5062-5069, 1995). Disruption of cpxR, which has been thought to be the cognate response regulator of cpxA (J. Dong, S. Iuchi, H.-S. Kwan, Z. Lue, and E. C. C. Lin, Gene 136:227-230, 1993), abolished virF expression almost completely. Purified CpxR bound directly to the upstream region of virF. Binding capacity was enhanced when CpxR was phosphorylated by coincubation with acetyl phosphate in vitro. Furthermore, we observed that phosphorylated CpxR could activate virF transcription in vitro. These results clearly indicated that CpxR was an essential activator for virF expression and strongly suggested that the binding of phosphorylated CpxR to the target site upstream of the virF gene induced a direct activation of virF transcription.
Figures

References
-
- Adler B, Sasakawa C, Tobe T, Makino S, Komatsu K, Yoshikawa M. A dual transcriptional activation system for the 230kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol. 1989;3:627–635. - PubMed
-
- Albright L M, Huala E, Ausubel F M. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu Rev Genet. 1989;23:311–336. - PubMed
-
- Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

