A region in the Bacillus subtilis transcription factor Spo0A that is important for spoIIG promoter activation
- PMID: 9658000
- PMCID: PMC107325
- DOI: 10.1128/JB.180.14.3578-3583.1998
A region in the Bacillus subtilis transcription factor Spo0A that is important for spoIIG promoter activation
Abstract
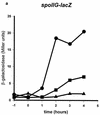
Spo0A is a DNA binding protein in Bacillus subtilis required for the activation of spoIIG and other promoters at the onset of endospore formation. Activation of some of these promoters may involve interaction of Spo0A and the sigmaA subunit of RNA polymerase. Previous studies identified two single-amino-acid substitutions in sigmaA, K356E and H359R, that specifically impaired Spo0A-dependent transcription in vivo. Here we report the identification of an amino acid substitution in Spo0A (S231F) that suppressed the sporulation deficiency due to the H359R substitution in sigmaA. We also found that the S231F substitution partially restored use of the spoIIG promoter by the sigmaA H359R RNA polymerase in vitro. Alanine substitutions in the 231 region of Spo0A revealed an additional amino acid residue important for spoIIG promoter activation, I229. This amino acid substitution in Spo0A did not affect repression of abrB transcription, indicating that the alanine-substituted Spo0A was not defective in DNA binding. Moreover, the alanine-substituted Spo0A protein activated the spoIIA promoter; therefore, this region of Spo0A is probably not required for Spo0A-dependent, sigmaH-directed transcription. These and other results suggest that the region of Spo0A near position 229 is involved in sigmaA-dependent promoter activation.
Figures
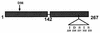




Similar articles
-
Alpha-helix E of Spo0A is required for sigmaA- but not for sigmaH-dependent promoter activation in Bacillus subtilis.J Bacteriol. 2004 Feb;186(4):1078-83. doi: 10.1128/JB.186.4.1078-1083.2004. J Bacteriol. 2004. PMID: 14762002 Free PMC article.
-
Activation of the Bacillus subtilis spoIIG promoter requires interaction of Spo0A and the sigma subunit of RNA polymerase.J Bacteriol. 1997 Sep;179(17):5605-8. doi: 10.1128/jb.179.17.5605-5608.1997. J Bacteriol. 1997. PMID: 9287022 Free PMC article.
-
A region in Bacillus subtilis sigmaH required for Spo0A-dependent promoter activity.J Bacteriol. 1998 Sep;180(18):4987-90. doi: 10.1128/JB.180.18.4987-4990.1998. J Bacteriol. 1998. PMID: 9733708 Free PMC article.
-
Surfaces of Spo0A and RNA polymerase sigma factor A that interact at the spoIIG promoter in Bacillus subtilis.J Bacteriol. 2004 Jan;186(1):200-6. doi: 10.1128/JB.186.1.200-206.2004. J Bacteriol. 2004. PMID: 14679239 Free PMC article.
-
ROMA: an in vitro approach to defining target genes for transcription regulators.Methods. 2009 Jan;47(1):73-7. doi: 10.1016/j.ymeth.2008.10.009. Epub 2008 Oct 21. Methods. 2009. PMID: 18948201 Free PMC article. Review.
Cited by
-
A gene encoding a holin-like protein involved in spore morphogenesis and spore germination in Bacillus subtilis.J Bacteriol. 2005 Sep;187(18):6443-53. doi: 10.1128/JB.187.18.6443-6453.2005. J Bacteriol. 2005. PMID: 16159778 Free PMC article.
-
Promoter activation by repositioning of RNA polymerase.J Bacteriol. 2008 May;190(9):3110-7. doi: 10.1128/JB.00096-08. Epub 2008 Feb 22. J Bacteriol. 2008. PMID: 18296515 Free PMC article.
-
Alpha-helix E of Spo0A is required for sigmaA- but not for sigmaH-dependent promoter activation in Bacillus subtilis.J Bacteriol. 2004 Feb;186(4):1078-83. doi: 10.1128/JB.186.4.1078-1083.2004. J Bacteriol. 2004. PMID: 14762002 Free PMC article.
-
Spo0A mutants of Bacillus subtilis with sigma factor-specific defects in transcription activation.J Bacteriol. 1998 Jul;180(14):3584-91. doi: 10.1128/JB.180.14.3584-3591.1998. J Bacteriol. 1998. PMID: 9658001 Free PMC article.
-
An A257V mutation in the bacillus subtilis response regulator Spo0A prevents regulated expression of promoters with low-consensus binding sites.J Bacteriol. 2009 Sep;191(17):5489-98. doi: 10.1128/JB.00590-09. Epub 2009 Jul 6. J Bacteriol. 2009. PMID: 19581368 Free PMC article.
References
-
- Baldus J M, Buckner C M, Moran C P., Jr Evidence that the transcriptional activator Spo0A interacts with two sigma factors in Bacillus subtilis. Mol Microbiol. 1995;17:281–290. - PubMed
-
- Bird T H, Grimsley J K, Hoch J A, Spiegelman G B. Phosphorylation of Spo0A activates its stimulation of in vitro transcription from the Bacillus subtilis spoIIG operon. Mol Microbiol. 1993;9:741–749. - PubMed
-
- Buckner, C. M., and C. P. Moran, Jr. 1998. Unpublished data.
-
- Busby S, Ebright R H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

