Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for L-idonic acid catabolism in Escherichia coli
- PMID: 9658018
- PMCID: PMC107343
- DOI: 10.1128/JB.180.14.3704-3710.1998
Sequence analysis of the GntII (subsidiary) system for gluconate metabolism reveals a novel pathway for L-idonic acid catabolism in Escherichia coli
Abstract
The presence of two systems in Escherichia coli for gluconate transport and phosphorylation is puzzling. The main system, GntI, is well characterized, while the subsidiary system, GntII, is poorly understood. Genomic sequence analysis of the region known to contain genes of the GntII system led to a hypothesis which was tested biochemically and confirmed: the GntII system encodes a pathway for catabolism of L-idonic acid in which D-gluconate is an intermediate. The genes have been named accordingly: the idnK gene, encoding a thermosensitive gluconate kinase, is monocistronic and transcribed divergently from the idnD-idnO-idnT-idnR operon, which encodes L-idonate 5-dehydrogenase, 5-keto-D-gluconate 5-reductase, an L-idonate transporter, and an L-idonate regulatory protein, respectively. The metabolic sequence is as follows: IdnT allows uptake of L-idonate; IdnD catalyzes a reversible oxidation of L-idonate to form 5-ketogluconate; IdnO catalyzes a reversible reduction of 5-ketogluconate to form D-gluconate; IdnK catalyzes an ATP-dependent phosphorylation of D-gluconate to form 6-phosphogluconate, which is metabolized further via the Entner-Doudoroff pathway; and IdnR appears to act as a positive regulator of the IdnR regulon, with L-idonate or 5-ketogluconate serving as the true inducer of the pathway. The L-idonate 5-dehydrogenase and 5-keto-D-gluconate 5-reductase reactions were characterized both chemically and biochemically by using crude cell extracts, and it was firmly established that these two enzymes allow for the redox-coupled interconversion of L-idonate and D-gluconate via the intermediate 5-ketogluconate. E. coli K-12 strains are able to utilize L-idonate as the sole carbon and energy source, and as predicted, the ability of idnD, idnK, idnR, and edd mutants to grow on L-idonate is altered.
Figures

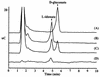
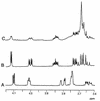
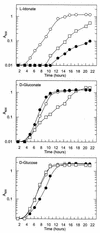
References
-
- Ashwell G. Enzymes of glucuronic and galacturonic acid metabolism in bacteria. Methods Enzymol. 1962;5:190–208.
-
- Bächi B, Kornberg H L. Genes involved in the uptake and catabolism of gluconate by Escherichia coli. J Gen Microbiol. 1975;90:321–335. - PubMed
-
- Bausch C, Peekhaus N, Blais T, Conway T. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Characterization of the gluconate subsidiary system (GntII) in Escherichia coli, abstr. K-75; p. 354.
-
- Bernaerts M, DeLey J. 2,5-Diketogluconate formation by Chromobacterium. Antonie Leeuwenhoek. 1971;37:185–195. - PubMed
-
- Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

