The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF
- PMID: 9658067
- PMCID: PMC109766
- DOI: 10.1128/JVI.72.8.6291-6297.1998
The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF
Abstract
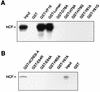
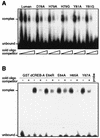
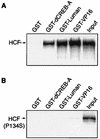
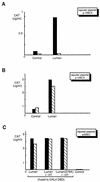
In human cells infected with herpes simplex virus (HSV), viral gene expression is initiated by the virion protein VP16. VP16 does not bind DNA directly but forms a multiprotein complex on the viral immediate-early gene promoters with two cellular proteins: the POU domain protein Oct-1 and host cell factor (HCF; also called C1, VCAF, and CFF). Despite its apparent role in stabilizing the VP16-induced transcription complex, the natural biological role of HCF is unclear. Only recently HCF has been implicated in control of the cell cycle. To determine the role of HCF in cells and answer why HSV has evolved an HCF-dependent mechanism for the initiation of the lytic cycle, we identified the first human ligand for HCF (R. Lu et al., Mol. Cell. Biol. 17:5117-5126, 1997). This protein, Luman, is a member of the CREB/ATF family of transcription factors that can activate transcription from promoters containing cyclic AMP response elements (CRE). Here we provide evidence that Luman and VP16 share two important structural features: an acidic activation domain and a common mechanism for binding HCF. We found that Luman, its homolog in Drosophila, dCREB-A (also known as BBF-2), and VP16 bind to HCF by a motif, (D/E)HXY(S/A), present in all three proteins. In addition, a mutation (P134S) in HCF that prevents VP16 binding also abolishes its binding to Luman and dCREB-A. We also show that while interaction with HCF is not required for the ability of Luman to activate transcription when tethered to the GAL4 promoter, it appears to be essential for Luman to activate transcription through CRE sites. These data suggest that the HCF-Luman interaction may represent a conserved mechanism for transcriptional regulation in metazoans, and HSV mimics this interaction with HCF to monitor the physiological state of the host cell.
Figures






References
-
- Abel T, Bhatt R, Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992;6:466–480. . (Erratum, 7:719, 1993.) - PubMed
-
- Burbelo P D, Gabriel G C, Kibbey M C, Yamada Y, Kleinman H K, Weeks B S. LZIP-1 and LZIP-2: two novel members of the bZIP family. Gene. 1994;139:241–245. - PubMed
-
- Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

