The FGF receptor-1 tyrosine kinase domain regulates myogenesis but is not sufficient to stimulate proliferation
- PMID: 9660877
- PMCID: PMC2133035
- DOI: 10.1083/jcb.142.1.241
The FGF receptor-1 tyrosine kinase domain regulates myogenesis but is not sufficient to stimulate proliferation
Abstract
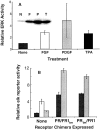
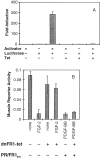
Ligand-stimulated activation of FGF receptors (FGFRs) in skeletal muscle cells represses terminal myogenic differentiation. Skeletal muscle cell lines and subsets of primary cells are dependent on FGFs to repress myogenesis and maintain growth. To understand the intracellular events that transduce these signals, MM14 skeletal muscle cells were transfected with expression vectors encoding chimeric receptors. The chimeras are comprised of the PDGF beta receptor (PDGFbetaR) extracellular domain, the FGFR-1 intracellular domain, and either the PDGFbetaR or FGFR-1 transmembrane domain. The chimeric receptors were autophosphorylated upon PDGF-BB stimulation and are capable of stimulating mitogen-activated protein kinase activity. Activation of the tyrosine kinase domain of either chimera repressed myogenesis, suggesting intracellular responses regulating skeletal muscle differentiation are transduced by activation of the FGFR-1 tyrosine kinase. Unexpectedly, we found that activation of either chimeric receptor failed to stimulate cellular proliferation. Thus, it appears that regulation of skeletal muscle differentiation by FGFs requires only activation of the FGFR tyrosine kinase. In contrast, stimulation of proliferation may require additional, as yet unidentified, signals involving the receptor ectodomain, the FGF ligand, and heparan sulfate either alone, or in combination.
Figures
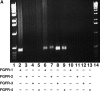

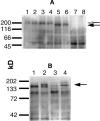
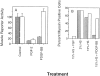
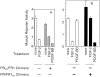




Similar articles
-
A requirement for fibroblast growth factor in regulation of skeletal muscle growth and differentiation cannot be replaced by activation of platelet-derived growth factor signaling pathways.Mol Cell Biol. 1995 Jun;15(6):3238-46. doi: 10.1128/MCB.15.6.3238. Mol Cell Biol. 1995. PMID: 7760819 Free PMC article.
-
Platelet-derived growth factor-BB-induced human smooth muscle cell proliferation depends on basic FGF release and FGFR-1 activation.Circ Res. 2005 Feb 4;96(2):172-9. doi: 10.1161/01.RES.0000154595.87608.db. Epub 2004 Dec 29. Circ Res. 2005. PMID: 15625285
-
Fibroblast growth factor receptor-1 mediates the inhibition of endothelial cell proliferation and the promotion of skeletal myoblast differentiation by SPARC: a role for protein kinase A.J Cell Biochem. 2003 Oct 1;90(2):408-23. doi: 10.1002/jcb.10645. J Cell Biochem. 2003. PMID: 14505356
-
Heparan sulfate fibroblast growth factor receptor complex: structure-function relationships.Mol Reprod Dev. 1994 Sep;39(1):69-81; discusison 81-2. doi: 10.1002/mrd.1080390112. Mol Reprod Dev. 1994. PMID: 7999363 Review.
-
Fibroblast growth factors and their receptors: an information network controlling tissue growth, morphogenesis and repair.Prog Growth Factor Res. 1994;5(4):353-77. doi: 10.1016/0955-2235(94)00007-8. Prog Growth Factor Res. 1994. PMID: 7780086 Review.
Cited by
-
Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination.J Cell Biol. 2007 Jun 4;177(5):769-79. doi: 10.1083/jcb.200608122. J Cell Biol. 2007. PMID: 17548510 Free PMC article.
-
Oncogenic Ras-induced proliferation requires autocrine fibroblast growth factor 2 signaling in skeletal muscle cells.J Cell Biol. 2001 Mar 19;152(6):1301-5. doi: 10.1083/jcb.152.6.1301. J Cell Biol. 2001. PMID: 11257129 Free PMC article.
-
Sarcopenia: Aging-Related Loss of Muscle Mass and Function.Physiol Rev. 2019 Jan 1;99(1):427-511. doi: 10.1152/physrev.00061.2017. Physiol Rev. 2019. PMID: 30427277 Free PMC article. Review.
-
FGF/FGFR signaling in health and disease.Signal Transduct Target Ther. 2020 Sep 2;5(1):181. doi: 10.1038/s41392-020-00222-7. Signal Transduct Target Ther. 2020. PMID: 32879300 Free PMC article. Review.
-
The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis.Skelet Muscle. 2011 Sep 8;1:29. doi: 10.1186/2044-5040-1-29. Skelet Muscle. 2011. PMID: 21902831 Free PMC article.
References
-
- Bargmann CI, Hung MC, Weinberg RA. Multiple independent activations of the neu oncogene by a point mutation altering the transmembrane domain of p185. Cell. 1986;45:649–57. - PubMed
-
- Burgess WH, Shaheen AM, Ravera M, Jaye M, Donohue PJ, Winkles JA. Possible dissociation of the heparin-binding and mitogenic activities of heparin-binding (acidic fibroblast) growth factor-1 from its receptor-binding activities by site-directed mutagenesis of a single lysine residue. J Cell Biol. 1990;111:2129–2138. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

