Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates
- PMID: 9665988
- PMCID: PMC105003
- DOI: 10.1128/JCM.36.8.2187-2190.1998
Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates
Abstract
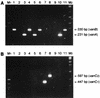
Enterococci with resistance to glycopeptides have recently emerged in Australia. We developed multiplex PCR assays for vanA, vanB, vanC1, and vanC2 or vanC3 in order to examine the genetic basis for vancomycin resistance in Australian isolates of vancomycin-resistant Enterococcus faecium and E. faecalis (VRE). The predominant genotype from human clinical E. faecium isolates was vanB. The PCR van genotype was consistent with the resistance phenotype in all but six cases. One vanA E. faecalis isolate had a VanB phenotype, one vanB E. faecium isolate had a VanA phenotype, and four E. faecalis isolates were consistently negative for vanA, vanB, vanC1, and vanC2 or vanC3, even though they exhibited a VanB phenotype. These four isolates were subsequently examined for the presence of vanD by published methods and were found to be negative. No vancomycin-susceptible strains produced a PCR product. On the basis of our findings the epidemiology of VRE in Australia appears to be different from that in either the United States or Europe. Our multiplex PCR assays gave a rapid and accurate method for determining the genotype and confirming the identification of glycopeptide-resistant enterococci. Rapid and accurate methods are essential, because laboratory-based surveillance is critical in programs for the detection, control, and prevention of the transmission of glycopeptide-resistant enterococci.
Figures
References
-
- Aarestrup F M, Ahrens P, Madsen M, Pallesen L V, Poulsen R L, Westh H. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob Agents Chemother. 1996;40:1938–1940. - PMC - PubMed
-
- AB Biodisk. Etest technical guide 3B. Solna, Sweden: AB Biodisk; 1995.
-
- Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. - PubMed
-
- Branley J, Yan B, Benn R A V. Vancomycin-resistant Enterococcus faecalis. Med J Aust. 1996;165:292. . (Letter.) - PubMed
-
- Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

