Genetic relationship between Cryptococcus neoformans var. neoformans strains of serotypes A and D
- PMID: 9665991
- PMCID: PMC105008
- DOI: 10.1128/JCM.36.8.2200-2204.1998
Genetic relationship between Cryptococcus neoformans var. neoformans strains of serotypes A and D
Abstract
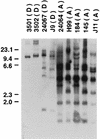
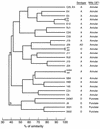
Cryptococcus neoformans serotypes A and D are responsible for the overwhelming majority of infections in patients with AIDS. The genetic relationship between the serotypes is poorly understood, but there are significant differences in the epidemiology and clinical presentation of serotype A and D infections. We evaluated the genetic relationship between reference C. neoformans strains belonging to serotypes A and D by analyzing their URA5 sequences and restriction fragment length polymorphisms (RFLPs) with the C. neoformans repetitive element 1 (CNRE-1) probe. The results were compared to those previously obtained for isolates from Brazil and New York City by the same typing methods, and dendrograms were generated. Serotype A and D strains produced distinct RFLP patterns consistent with their separation into two major clusters in the dendrogram generated on the basis of RFLP data. Similarly, serotype A and D strains clustered independently of the basis of the nucleotide sequences of their URA5 genes. Pairwise comparisons revealed average numbers of nucleotide differences within serotypes A and D of 3.0 +/- 1.7 and 7.2 +/- 3.4, respectively (P < 0.0001), and between serotypes A and D of 41.9 +/- 2.7. In summary, our results indicate phylogenetic differences between the two serotypes of C. neoformans var. neoformans and suggest that these serotypes could probably be considered different varieties of C. neoformans.
Figures


References
-
- Aulakh H S, Straus S E, Kwon-Chung K J. Genetic relatedness of Filobasidiella neoformans (Cryptococcus neoformans) and Filobasidiella bacillispora (Cryptococcus bacillispora) as determined by DNA base composition and sequence homology studies. Int J Syst Bacteriol. 1981;31:97–103.
-
- Bennett J E, Kwon-Chung K J, Howard D. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1984;105:582–586. - PubMed
-
- Boekhout T, van Belkum A, Leenders A C, Verbrugh H A, Mukamurangwa P, Swinne D, Scheffers W A. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

