Exonic sequences in the 5' untranslated region of alpha-tubulin mRNA modulate trans splicing in Trypanosoma brucei
- PMID: 9671472
- PMCID: PMC109048
- DOI: 10.1128/MCB.18.8.4620
Exonic sequences in the 5' untranslated region of alpha-tubulin mRNA modulate trans splicing in Trypanosoma brucei
Abstract
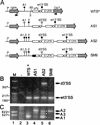
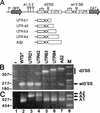
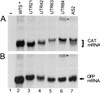
Previous studies have identified a conserved AG dinucleotide at the 3' splice site (3'SS) and a polypyrimidine (pPy) tract that are required for trans splicing of polycistronic pre-mRNAs in trypanosomatids. Furthermore, the pPy tract of the Trypanosoma brucei alpha-tubulin 3'SS region is required to specify accurate 3'-end formation of the upstream beta-tubulin gene and trans splicing of the downstream alpha-tubulin gene. Here, we employed an in vivo cis competition assay to determine whether sequences other than those of the AG dinucleotide and the pPy tract were required for 3'SS identification. Our results indicate that a minimal alpha-tubulin 3'SS, from the putative branch site region to the AG dinucleotide, is not sufficient for recognition by the trans-splicing machinery and that polyadenylation is strictly dependent on downstream trans splicing. We show that efficient use of the alpha-tubulin 3'SS is dependent upon the presence of exon sequences. Furthermore, beta-tubulin, but not actin exon sequences or unrelated plasmid sequences, can replace alpha-tubulin exon sequences for accurate trans-splice-site selection. Taken together, these results support a model in which the informational content required for efficient trans splicing of the alpha-tubulin pre-mRNA includes exon sequences which are involved in modulation of trans-splicing efficiency. Sequences that positively regulate trans splicing might be similar to cis-splicing enhancers described in other systems.
Figures




Similar articles
-
Systematic study of sequence motifs for RNA trans splicing in Trypanosoma brucei.Mol Cell Biol. 2005 Nov;25(21):9586-94. doi: 10.1128/MCB.25.21.9586-9594.2005. Mol Cell Biol. 2005. PMID: 16227607 Free PMC article.
-
A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes.Genes Dev. 1994 Feb 15;8(4):491-501. doi: 10.1101/gad.8.4.491. Genes Dev. 1994. PMID: 7907303
-
A suboptimal 5' splice site downstream of HIV-1 splice site A1 is required for unspliced viral mRNA accumulation and efficient virus replication.Retrovirology. 2006 Feb 3;3:10. doi: 10.1186/1742-4690-3-10. Retrovirology. 2006. PMID: 16457729 Free PMC article.
-
Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts.Mol Cell Biol. 1993 Jan;13(1):720-5. doi: 10.1128/mcb.13.1.720-725.1993. Mol Cell Biol. 1993. PMID: 8417363 Free PMC article.
-
Splicing Enhancers at Intron-Exon Borders Participate in Acceptor Splice Sites Recognition.Int J Mol Sci. 2020 Sep 8;21(18):6553. doi: 10.3390/ijms21186553. Int J Mol Sci. 2020. PMID: 32911621 Free PMC article.
Cited by
-
The short interspersed repetitive element of Trypanosoma cruzi, SIRE, is part of VIPER, an unusual retroelement related to long terminal repeat retrotransposons.Proc Natl Acad Sci U S A. 2000 Feb 29;97(5):2128-33. doi: 10.1073/pnas.050578397. Proc Natl Acad Sci U S A. 2000. PMID: 10688909 Free PMC article.
-
The Trypanosoma brucei RNA-binding protein DRBD18 ensures correct mRNA trans splicing and polyadenylation patterns.RNA. 2022 Sep;28(9):1239-1262. doi: 10.1261/rna.079258.122. Epub 2022 Jul 6. RNA. 2022. PMID: 35793904 Free PMC article.
-
Different trans RNA splicing events in bloodstream and procyclic Trypanosoma brucei.Mol Biochem Parasitol. 2008 Jun;159(2):134-7. doi: 10.1016/j.molbiopara.2008.02.006. Epub 2008 Feb 15. Mol Biochem Parasitol. 2008. PMID: 18384893 Free PMC article.
-
Establishment of an in vitro trans-splicing system in Trypanosoma brucei that requires endogenous spliced leader RNA.Nucleic Acids Res. 2010 Jun;38(10):e114. doi: 10.1093/nar/gkq065. Epub 2010 Feb 16. Nucleic Acids Res. 2010. PMID: 20159996 Free PMC article.
-
3' splice site recognition in nematode trans-splicing involves enhancer-dependent recruitment of U2 snRNP.RNA. 2001 Jun;7(6):785-92. doi: 10.1017/s1355838201010263. RNA. 2001. PMID: 11421357 Free PMC article.
References
-
- Chiara M D, Reed R. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature. 1995;375:510–513. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
