Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences
- PMID: 9671477
- PMCID: PMC109053
- DOI: 10.1128/MCB.18.8.4670
Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences
Abstract
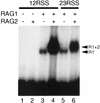
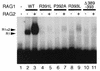
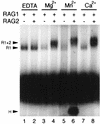
The RAG1 and RAG2 proteins initiate V(D)J recombination by introducing double-strand breaks at the border between a recombination signal sequence (RSS) and a coding segment. To understand the distinct functions of RAG1 and RAG2 in signal recognition, we have compared the DNA binding activities of RAG1 alone and RAG1 plus RAG2 by gel retardation and footprinting analyses. RAG1 exhibits only a three- to fivefold preference for binding DNA containing an RSS over random sequence DNA. Although direct binding of RAG2 by itself was not detected, the presence of both RAG1 and RAG2 results in the formation of a RAG1-RAG2-DNA complex which is more stable and more specific than the RAG1-DNA complex and is active in V(D)J cleavage. These results suggest that biologically effective discrimination between an RSS and nonspecific sequences requires both RAG1 and RAG2. Unlike the binding of RAG1 plus RAG2, RAG1 can bind to DNA in the absence of a divalent metal ion and does not require the presence of coding flank sequence. Footprinting of the RAG1-RAG2 complex with 1, 10-phenanthroline-copper and dimethyl sulfate protection reveal that both the heptamer and the nonamer are involved. The nonamer is protected, with extensive protein contacts within the minor groove. Conversely, the heptamer is rendered more accessible to chemical attack, suggesting that binding of RAG1 plus RAG2 distorts the DNA near the coding/signal border.
Figures





References
-
- Akamatsu Y, Tsurushita N, Nagawa F, Matsuoka M, Okazaki K, Imai M, Sakano H. Essential residues in V(D)J recombination signals. J Immunol. 1994;153:4520–4529. - PubMed
-
- Akamatsu, Y. Unpublished observations.
-
- Akira S, Okazaki K, Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987;238:1134–1138. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1989.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
