Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in chromatin assembly factor I and histone regulatory proteins
- PMID: 9671488
- PMCID: PMC109064
- DOI: 10.1128/MCB.18.8.4783
Yeast Ty1 retrotransposition is stimulated by a synergistic interaction between mutations in chromatin assembly factor I and histone regulatory proteins
Abstract
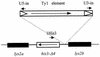
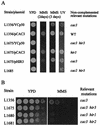
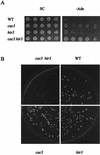
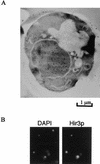
A screen for host mutations which increase the rate of transposition of Ty1 and Ty2 into a chromosomal target was used to identify factors influencing retroelement transposition. The fortuitous presence of a mutation in the CAC3 gene in the strain in which this screen was undertaken enabled us to discover that double mutaions of cac3 and hir3, but neither of the two single mutations, caused a dramatic increase in the rate of retrotransposition. We further showed that this effect was not due to an increase in the overall level of Ty1 mRNA. Two subtle cac3 phenotypes, slight methyl methanesulfonate (MMS) sensitivity and reduction of telomeric silencing, were significantly enhanced in the cac3 hir3 double mutant. In addition, the growth rate of the double mutant was reduced. HIR3 belongs to a class of HIR genes that regulate the transcription of histones, while Cac3p, together with Cac1p and Cac2p, forms chromatin assembly factor I. Other combinations of mutations in cac and hir genes (cac3 hir1, cac3 hir2, and cac2 hir3) also increase Ty transposition and MMS sensitivity and reduce the growth rate. A model explaining the synergistic interaction between cac and hir mutations in terms of alterations in chromatin structure is proposed.
Figures


Similar articles
-
Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing.Mol Cell Biol. 2002 Jan;22(2):614-25. doi: 10.1128/MCB.22.2.614-625.2002. Mol Cell Biol. 2002. PMID: 11756556 Free PMC article.
-
Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing.Curr Biol. 2001 Apr 3;11(7):463-73. doi: 10.1016/s0960-9822(01)00140-3. Curr Biol. 2001. PMID: 11412995
-
Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure.Genetics. 2002 Dec;162(4):1557-71. doi: 10.1093/genetics/162.4.1557. Genetics. 2002. PMID: 12524332 Free PMC article.
-
Chromatin assembly: the kinetochore connection.Curr Biol. 2002 Apr 2;12(7):R256-8. doi: 10.1016/s0960-9822(02)00786-8. Curr Biol. 2002. PMID: 11937044 Review.
-
Happy together: the life and times of Ty retrotransposons and their hosts.Cytogenet Genome Res. 2005;110(1-4):70-90. doi: 10.1159/000084940. Cytogenet Genome Res. 2005. PMID: 16093660 Review.
Cited by
-
The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF.Genes Dev. 2005 Nov 1;19(21):2534-9. doi: 10.1101/gad.1341105. Genes Dev. 2005. PMID: 16264190 Free PMC article.
-
Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus.Proc Natl Acad Sci U S A. 2010 Oct 26;107(43):18551-6. doi: 10.1073/pnas.1014023107. Epub 2010 Oct 11. Proc Natl Acad Sci U S A. 2010. PMID: 20937885 Free PMC article.
-
A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage.Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16934-9. doi: 10.1073/pnas.262669299. Epub 2002 Dec 13. Proc Natl Acad Sci U S A. 2002. PMID: 12482937 Free PMC article.
-
Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription.Genetics. 2001 Jun;158(2):587-96. doi: 10.1093/genetics/158.2.587. Genetics. 2001. PMID: 11404324 Free PMC article.
-
A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae.Genetics. 1999 Jul;152(3):921-32. doi: 10.1093/genetics/152.3.921. Genetics. 1999. PMID: 10388812 Free PMC article.
References
-
- Boeke J D, Garfinkel D J, Styles C A, Fink G R. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. - PubMed
-
- Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
