Cloning and functional expression of an Aplysia 5-HT receptor negatively coupled to adenylate cyclase
- PMID: 9671650
- PMCID: PMC6793050
- DOI: 10.1523/JNEUROSCI.18-15-05586.1998
Cloning and functional expression of an Aplysia 5-HT receptor negatively coupled to adenylate cyclase
Abstract
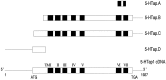
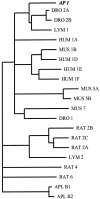
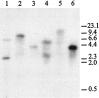
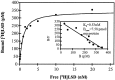
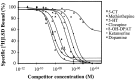
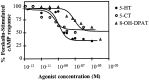
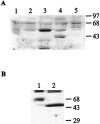
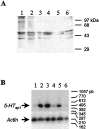
Serotonin (5-HT) is involved in the control of various behaviors in Aplysia californica, including reproduction, feeding, locomotion, circadian rhythm, synaptic plasticity, and synaptic growth. The large variety of functions of 5-HT is mediated by different receptor subtypes that are coupled to different second-messenger systems. Here, we report the cloning of a cDNA coding for an Aplysia G-protein-coupled 5-HT receptor (5-HTap1). Its deduced amino acid sequence resembles those of the 5-HT1 receptor subfamily. When expressed in stable cell lines, 5-HTap1 exhibits high-affinity binding for the serotonergic radioligand [N-methyl-3H]lysergic acid diethylamide. This binding is competed by several 5-HT agonists and antagonists, and the pharmacological profile of inhibition has some similarities with those of 5-HT1 and 5-HT7 receptors. Application of 5-HT or its agonists 5-carboxamidotryptamine maleate and (+/-)-8-hydroxy-2-(di-n-propyl-amino) tetralin hydrobromide on cells transformed with 5-HTap1 produced a dose-dependent inhibition of forskolin-stimulated cAMP accumulation. 5-HTap1 is thus negatively coupled to adenylate cyclase. The production of antiserum against the 5-HTap1 receptor allowed us to examine its expression in animal tissues. The receptor protein is detected in every tissue examined, although it seems only weakly expressed in some samples. The receptor is also found in every ganglia of the nervous system, both in the sheath and in the neurons. 5-HTap1 mRNA is absent from the sheath, indicating that the protein observed there is probably located on the nerve terminals.
Figures

References
-
- Aloyz RS, DesGroseillers L. Processing of the L5–67 precursor peptide and characterization of LUQIN in the LUQ neurons of Aplysia californica. Peptides. 1995;16:331–338. - PubMed
-
- Andrade R, Malenka RC, Nicall RA. A G-protein couples serotonin and GABAB receptors to the same channels in hippocampus. Science. 1986;234:1261–1265. - PubMed
-
- Angers A, DesGroseillers L. Alternative splicing and genomic organization of the L567 gene of Aplysia californica. Gene. 1998;208:271–277. - PubMed
-
- Ansanay H, Sebben M, Bockaert J, Dumuis A. Characterization of homologous 5-hydroxytryptamine4 receptor desensitization in colliculi neurons. Mol Pharmacol. 1992;42:808–816. - PubMed
-
- Bacskai BJ, Hochner B, Mahaut-Smith M, Adams SR, Kaang BK, Kandel ER, Tsien RY. Spatially resolved dynamics of cAMP and protein kinase A subunits in Aplysia sensory neurons. Science. 1993;260:222–226. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
