3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase
- PMID: 9671708
- PMCID: PMC21106
- DOI: 10.1073/pnas.95.15.8508
3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase
Abstract
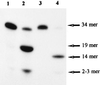
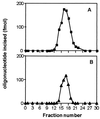
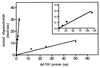
Exocyclic DNA adducts are generated in cellular DNA by various industrial pollutants such as the carcinogen vinyl chloride and by endogenous products of lipid peroxidation. The etheno derivatives of purine and pyrimidine bases 3,N4-ethenocytosine (epsilonC), 1, N6-ethenoadenine (epsilonA), N2,3-ethenoguanine, and 1, N2-ethenoguanine cause mutations. The epsilonA residues are excised by the human and the Escherichia coli 3-methyladenine-DNA glycosylases (ANPG and AlkA proteins, respectively), but the enzymes repairing epsilonC residues have not yet been described. We have identified two homologous proteins present in human cells and E. coli that remove epsilonC residues by a DNA glycosylase activity. The human enzyme is an activity of the mismatch-specific thymine-DNA glycosylase (hTDG). The bacterial enzyme is the double-stranded uracil-DNA glycosylase (dsUDG) that is the homologue of the hTDG. In addition to uracil and epsilonC-DNA glycosylase activity, the dsUDG protein repairs thymine in a G/T mismatch. The fact that epsilonC is recognized and efficiently excised by the E. coli dsUDG and hTDG proteins in vitro suggests that these enzymes may be responsible for the repair of this mutagenic lesion in vivo and be important contributors to genetic stability.
Figures


Similar articles
-
Enzymology of the repair of etheno adducts in mammalian cells and in Escherichia coli.IARC Sci Publ. 1999;(150):249-61. IARC Sci Publ. 1999. PMID: 10626225
-
Novel activity of Escherichia coli mismatch uracil-DNA glycosylase (Mug) excising 8-(hydroxymethyl)-3,N4-ethenocytosine, a potential product resulting from glycidaldehyde reaction.Biochemistry. 2002 Feb 19;41(7):2158-65. doi: 10.1021/bi011542b. Biochemistry. 2002. PMID: 11841206
-
Chloroethylnitrosourea-derived ethano cytosine and adenine adducts are substrates for Escherichia coli glycosylases excising analogous etheno adducts.DNA Repair (Amst). 2004 Oct 5;3(10):1311-21. doi: 10.1016/j.dnarep.2004.04.015. DNA Repair (Amst). 2004. PMID: 15336626
-
Structure and function in the uracil-DNA glycosylase superfamily.Mutat Res. 2000 Aug 30;460(3-4):165-81. doi: 10.1016/s0921-8777(00)00025-2. Mutat Res. 2000. PMID: 10946227 Review.
-
Enzymology of repair of etheno-adducts.Mutat Res. 2003 Oct 29;531(1-2):219-29. doi: 10.1016/j.mrfmmm.2003.07.008. Mutat Res. 2003. PMID: 14637257 Review.
Cited by
-
Differential modes of DNA binding by mismatch uracil DNA glycosylase from Escherichia coli: implications for abasic lesion processing and enzyme communication in the base excision repair pathway.Nucleic Acids Res. 2011 Apr;39(7):2593-603. doi: 10.1093/nar/gkq913. Epub 2010 Nov 25. Nucleic Acids Res. 2011. PMID: 21112870 Free PMC article.
-
Direct repair of the exocyclic DNA adduct 1,N6-ethenoadenine by the DNA repair AlkB proteins.J Am Chem Soc. 2005 Oct 26;127(42):14594-5. doi: 10.1021/ja055957m. J Am Chem Soc. 2005. PMID: 16231911 Free PMC article.
-
Recent advances in the structural mechanisms of DNA glycosylases.Biochim Biophys Acta. 2013 Jan;1834(1):247-71. doi: 10.1016/j.bbapap.2012.10.005. Epub 2012 Oct 14. Biochim Biophys Acta. 2013. PMID: 23076011 Free PMC article. Review.
-
Repair of DNA Alkylation Damage by the Escherichia coli Adaptive Response Protein AlkB as Studied by ESI-TOF Mass Spectrometry.J Nucleic Acids. 2010 Oct 27;2010:369434. doi: 10.4061/2010/369434. J Nucleic Acids. 2010. PMID: 21048928 Free PMC article.
-
Aberrant repair initiated by mismatch-specific thymine-DNA glycosylases provides a mechanism for the mutational bias observed in CpG islands.Nucleic Acids Res. 2014 Jun;42(10):6300-13. doi: 10.1093/nar/gku246. Epub 2014 Apr 1. Nucleic Acids Res. 2014. PMID: 24692658 Free PMC article.
References
-
- Green T, Hathway D E. Chem Biol Interact. 1978;22:211–224. - PubMed
-
- Guengerich F P, Kim D-H. Chem Res Toxicol. 1991;4:413–421. - PubMed
-
- Dahl G A, Miller J A, Miller E C. Cancer Res. 1978;38:3793–3804. - PubMed
-
- Leithäuser M T, Liem A, Steward B C, Miller E C, Miller J A. Carcinogenesis. 1990;11:463–473. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

