Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases
- PMID: 9671747
- PMCID: PMC21145
- DOI: 10.1073/pnas.95.15.8733
Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases
Abstract
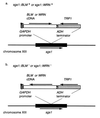
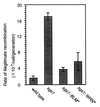
Bloom's syndrome (BS) and Werner's syndrome (WS) are genetic disorders in which an increased rate of chromosomal aberration is detected. The genes responsible for these diseases, BLM and WRN, have been found to be homologs of Escherichia coli recQ and Saccharomyces cerevisiae SGS1 genes. Here we show that yeast Sgs1 helicase acts as a suppressor of illegitimate recombination through homologous recombination and that human BLM and WRN helicases can suppress the increased homologous and illegitimate recombinations in the S. cerevisiae sgs1 mutant. The results imply a role of BLM and WRN helicases to control genomic stability in human cells. Similar to Sgs1 helicase, BLM helicase suppressed the cell growth in the top3 sgs1 mutation background and restored the increased sensitivity of the sgs1 mutant to hydroxyurea, but the WRN helicase did not. We discussed differential roles of BLM and WRN helicases in human cells. BLM- and WRN-bearing yeasts provide new useful models to investigate human BS and WS diseases.
Figures
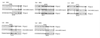



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

