Nitrilase and Fhit homologs are encoded as fusion proteins in Drosophila melanogaster and Caenorhabditis elegans
- PMID: 9671749
- PMCID: PMC21147
- DOI: 10.1073/pnas.95.15.8744
Nitrilase and Fhit homologs are encoded as fusion proteins in Drosophila melanogaster and Caenorhabditis elegans
Abstract
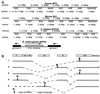
The tumor suppressor gene FHIT encompasses the common human chromosomal fragile site at 3p14.2 and numerous cancer cell biallelic deletions. To study Fhit function we cloned and characterized FHIT genes from Drosophila melanogaster and Caenorhabditis elegans. Both genes code for fusion proteins in which the Fhit domain is fused with a novel domain showing homology to bacterial and plant nitrilases; the D. melanogaster fusion protein exhibited diadenosine triphosphate (ApppA) hydrolase activity expected of an authentic Fhit homolog. In human and mouse, the nitrilase homologs and Fhit are encoded by two different genes: FHIT and NIT1, localized on chromosomes 3 and 1 in human, and 14 and 1 in mouse, respectively. We cloned and characterized human and murine NIT1 genes and determined their exon-intron structure, patterns of expression, and alternative processing of their mRNAs. The tissue specificity of expression of murine Fhit and Nit1 genes was nearly identical. Because fusion proteins with dual or triple enzymatic activities have been found to carry out specific steps in a given biochemical or biosynthetic pathway, we postulate that Fhit and Nit1 likewise collaborate in a biochemical or cellular pathway in mammalian cells.
Figures

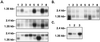


Similar articles
-
The Rosetta Stone Hypothesis-Based Interaction of the Tumor Suppressor Proteins Nit1 and Fhit.Cells. 2023 Jan 17;12(3):353. doi: 10.3390/cells12030353. Cells. 2023. PMID: 36766695 Free PMC article.
-
Biological functions of mammalian Nit1, the counterpart of the invertebrate NitFhit Rosetta stone protein, a possible tumor suppressor.J Biol Chem. 2006 Sep 22;281(38):28244-53. doi: 10.1074/jbc.M603590200. Epub 2006 Jul 24. J Biol Chem. 2006. PMID: 16864578
-
Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers.Curr Biol. 2000 Jul 27-Aug 10;10(15):907-17. doi: 10.1016/s0960-9822(00)00621-7. Curr Biol. 2000. PMID: 10959838
-
Hits, Fhits and Nits: beyond enzymatic function.Adv Enzyme Regul. 2011;51(1):208-17. doi: 10.1016/j.advenzreg.2010.09.003. Epub 2010 Oct 28. Adv Enzyme Regul. 2011. PMID: 21035495 Free PMC article. Review.
-
The histidine triad superfamily of nucleotide-binding proteins.J Cell Physiol. 1999 Nov;181(2):179-87. doi: 10.1002/(SICI)1097-4652(199911)181:2<179::AID-JCP1>3.0.CO;2-8. J Cell Physiol. 1999. PMID: 10497298 Free PMC article. Review.
Cited by
-
The Rosetta Stone Hypothesis-Based Interaction of the Tumor Suppressor Proteins Nit1 and Fhit.Cells. 2023 Jan 17;12(3):353. doi: 10.3390/cells12030353. Cells. 2023. PMID: 36766695 Free PMC article.
-
Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases.Biochemistry. 2002 Jul 23;41(29):9003-14. doi: 10.1021/bi025942q. Biochemistry. 2002. PMID: 12119013 Free PMC article.
-
Fhit-nucleotide specificity probed with novel fluorescent and fluorogenic substrates.J Biol Chem. 2000 Feb 18;275(7):4555-60. doi: 10.1074/jbc.275.7.4555. J Biol Chem. 2000. PMID: 10671479 Free PMC article.
-
Domain fusion analysis by applying relational algebra to protein sequence and domain databases.BMC Bioinformatics. 2003 May 6;4:16. doi: 10.1186/1471-2105-4-16. Epub 2003 May 6. BMC Bioinformatics. 2003. PMID: 12734020 Free PMC article.
-
Mammalian nitrilase 1 homologue Nit1 is a negative regulator in T cells.Int Immunol. 2009 Jun;21(6):691-703. doi: 10.1093/intimm/dxp038. Epub 2009 Apr 24. Int Immunol. 2009. PMID: 19395373 Free PMC article.
References
-
- Ohta M, Inoue H, Cotticelli M G, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. Cell. 1996;84:587–597. - PubMed
-
- Sozzi G, Veronese M L, Negrini M, Baffa R, Cotticelli M G, Inoue H, Tornielli S, Pilotti S, DeGregorio L, Pastorino V, et al. Cell. 1996;85:17–26. - PubMed
-
- Druck T, Hadaczek P, Fu T-B, Ohta M, Siprashvili Z, Baffa R, Negrini M, Kastury K, Veronese M L, Rosen D, et al. Cancer Res. 1997;57:504–512. - PubMed
-
- Panagopoulos I, Thelin S, Mertens F, Mitelman F, Aman P. Genes Chromosomes Cancer. 1997;19:215–219. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

