hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer
- PMID: 9671765
- PMCID: PMC21163
- DOI: 10.1073/pnas.95.15.8835
hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer
Abstract
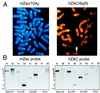
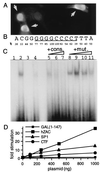
We previously reported the identification of mZac, a novel mouse zinc finger protein that shared with p53 the ability to regulate concomitantly apoptosis and cell cycle progression. We describe here the isolation, chromosomal localization, and functional in vitro characterization of its human homolog. hZAC is a widely expressed zinc finger protein that reveals transactivation and DNA-binding activity. hZAC inhibits tumor cell growth through induction of apoptotic cell death and G1 arrest. Thus hZAC, like its mouse counterpart, displays antiproliferative properties through pathways known to be central to the activity of p53. We mapped hZAC on chromosome 6q24-q25, a region frequently deleted in many solid tumors. Indeed, allelic loss at 6q24-q25 has been shown in breast and ovary cancers, melanomas, astrocytomas, and renal cell carcinomas. Furthermore, Abdollahi et al. [Abdollahi, A., Godwin, A. K., Miller, P. D., Getts, L. A., Schultz, D. C., Tagushi, T., Testa, J. R. & Hamilton, T. C. (1997) Cancer Res. 57, 2029-2034] recently isolated ZAC through its loss of expression in a surface epithelial ovary tumor model and accordingly named it Lot for "lost on transformation." In view of these observations, the functional properties we report here provide further arguments to consider hZAC as a tumor suppressor gene candidate.
Figures





References
-
- Hartwell L H, Kastan M B. Science. 1994;266:1821–1828. - PubMed
-
- Hussussian C J, Struewing J P, Goldstein A M, Higgins P A, Ally D S, Sheahan M D, Clark W H J, Tucker M A, Dracopoli N C. Nat Genet. 1994;8:15–21. - PubMed
-
- Chandrasekharappa S C, Guru S C, Manickam P, Olufemi S E, Collins F S, Emmert-Buck M R, Debelenko L V, Zhuang Z, Lubensky I A, Liotta L A, et al. Science. 1997;276:404–407. - PubMed
-
- Faienza M F, della Ragione F, Basso G, Coppola B, Miraglia del Giudice E, Schettini F, Iolascon A. Br J Haematol. 1996;93:632–636. - PubMed
-
- Mollenhauer J, Wiemann S, Scheurlen W, Korn B, Hayashi Y, Wilgenbus K K, von Deimling A, Poutska A. Nat Genet. 1997;17:32–39. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

