Characterization of the thermal stress response of Campylobacter jejuni
- PMID: 9673247
- PMCID: PMC108400
- DOI: 10.1128/IAI.66.8.3666-3672.1998
Characterization of the thermal stress response of Campylobacter jejuni
Abstract
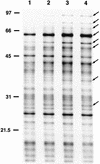
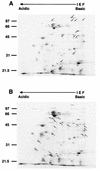
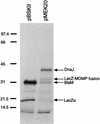
Campylobacter jejuni, a microaerophilic, gram-negative bacterium, is a common cause of gastrointestinal disease in humans. Heat shock proteins are a group of highly conserved, coregulated proteins that play important roles in enabling organisms to cope with physiological stresses. The primary aim of this study was to characterize the heat shock response of C. jejuni. Twenty-four proteins were preferentially synthesized by C. jejuni immediately following heat shock. Upon immunoscreening of Escherichia coli transformants harboring a Campylobacter genomic DNA library, one recombinant plasmid that encoded a heat shock protein was isolated. The recombinant plasmid, designated pMEK20, contained an open reading frame of 1,119 bp that was capable of encoding a protein of 372 amino acids with a calculated molecular mass of 41,436 Da. The deduced amino acid sequence of the open reading frame shared similarity with that of DnaJ, which belongs to the Hsp-40 family of molecular chaperones, from a number of bacteria. An E. coli dnaJ mutant was successfully complemented with the pMEK20 recombinant plasmid, as judged by the ability of bacteriophage lambda to form plaques, indicating that the C. jejuni gene encoding the 41-kDa protein is a functional homolog of the dnaJ gene from E. coli. The ability of each of two C. jejuni dnaJ mutants to form colonies at 46 degreesC was severely retarded, indicating that DnaJ plays an important role in C. jejuni thermotolerance. Experiments revealed that a C. jejuni DnaJ mutant was unable to colonize newly hatched Leghorn chickens, suggesting that heat shock proteins play a role in vivo.
Figures


References
-
- Bardwell J C A, Tilly K, Craig E, King J, Zylicz M, Georgopoulos C. The nucleotide sequence of the Escherichia coli K12 dnaJ+ gene: a gene that encodes a heat shock protein. J Biol Chem. 1986;261:1782–1785. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

