Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins
- PMID: 9679132
- PMCID: PMC2133057
- DOI: 10.1083/jcb.142.2.307
Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins
Abstract
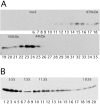
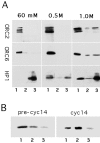
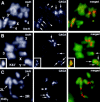
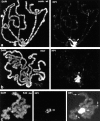
The distinct structural properties of heterochromatin accommodate a diverse group of vital chromosome functions, yet we have only rudimentary molecular details of its structure. A powerful tool in the analyses of its structure in Drosophila has been a group of mutations that reverse the repressive effect of heterochromatin on the expression of a gene placed next to it ectopically. Several genes from this group are known to encode proteins enriched in heterochromatin. The best characterized of these is the heterochromatin-associated protein, HP1. HP1 has no known DNA-binding activity, hence its incorporation into heterochromatin is likely to be dependent upon other proteins. To examine HP1 interacting proteins, we isolated three distinct oligomeric species of HP1 from the cytoplasm of early Drosophila embryos and analyzed their compositions. The two larger oligomers share two properties with the fraction of HP1 that is most tightly associated with the chromatin of interphase nuclei: an underphosphorylated HP1 isoform profile and an association with subunits of the origin recognition complex (ORC). We also found that HP1 localization into heterochromatin is disrupted in mutants for the ORC2 subunit. These findings support a role for the ORC-containing oligomers in localizing HP1 into Drosophila heterochromatin that is strikingly similar to the role of ORC in recruiting the Sir1 protein to silencing nucleation sites in Saccharomyces cerevisiae.
Figures




Similar articles
-
Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing.Mol Biol Cell. 2001 Jun;12(6):1671-85. doi: 10.1091/mbc.12.6.1671. Mol Biol Cell. 2001. PMID: 11408576 Free PMC article.
-
Mutations in the heterochromatin protein 1 (HP1) hinge domain affect HP1 protein interactions and chromosomal distribution.Chromosoma. 2005 Feb;113(7):370-84. doi: 10.1007/s00412-004-0324-2. Epub 2004 Dec 9. Chromosoma. 2005. PMID: 15592864
-
HP1/ORC complex and heterochromatin assembly.Genetica. 2003 Mar;117(2-3):127-34. doi: 10.1023/a:1022963223220. Genetica. 2003. PMID: 12723692
-
The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription.Bioessays. 2011 Apr;33(4):280-9. doi: 10.1002/bies.201000138. Epub 2011 Jan 27. Bioessays. 2011. PMID: 21271610 Review.
-
HP1 complexes and heterochromatin assembly.Curr Top Microbiol Immunol. 2003;274:53-77. doi: 10.1007/978-3-642-55747-7_3. Curr Top Microbiol Immunol. 2003. PMID: 12596904 Review.
Cited by
-
Linking DNA replication to heterochromatin silencing and epigenetic inheritance.Acta Biochim Biophys Sin (Shanghai). 2012 Jan;44(1):3-13. doi: 10.1093/abbs/gmr107. Acta Biochim Biophys Sin (Shanghai). 2012. PMID: 22194009 Free PMC article. Review.
-
Chromatin architectural proteins.Chromosome Res. 2006;14(1):39-51. doi: 10.1007/s10577-006-1025-x. Chromosome Res. 2006. PMID: 16506095 Review.
-
ORChestra coordinates the replication and repair music.Bioessays. 2023 Apr;45(4):e2200229. doi: 10.1002/bies.202200229. Epub 2023 Feb 22. Bioessays. 2023. PMID: 36811379 Free PMC article.
-
High- and low-mobility populations of HP1 in heterochromatin of mammalian cells.Mol Biol Cell. 2004 Jun;15(6):2819-33. doi: 10.1091/mbc.e03-11-0827. Epub 2004 Apr 2. Mol Biol Cell. 2004. PMID: 15064352 Free PMC article.
-
Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance.EMBO J. 2004 Jul 7;23(13):2651-63. doi: 10.1038/sj.emboj.7600255. Epub 2004 Jun 24. EMBO J. 2004. PMID: 15215892 Free PMC article.
References
-
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. - PubMed
-
- Baska K, Morawietz H, Dombrádi V, Axton M, Taubert H, Szabó G, Török I, Udvardy A, Gyurkovics H, Szöör B, Glover D, Reuter G, Gausz J. Mutations in the protein phosphatase 1 gene at 87B can differentially affect suppression of position-effect variegation and mitosis in Drosophila melanogaster. . Genetics. 1993;135:117–125. - PMC - PubMed
-
- Bell SP, Stillman B. ATP-dependent recognition of eucaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
-
- Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. - PubMed
-
- Bhat KM, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. The GAGA factor is required in the early Drosophilaembryo not only for transcriptional regulation but also for nuclear division. Development (Camb) 1996;22:1113–1124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

