The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded beta-sheet proteins by its sequence and properties
- PMID: 9683466
- PMCID: PMC107353
- DOI: 10.1128/JB.180.15.3741-3749.1998
The regulated outer membrane protein Omp21 from Comamonas acidovorans is identified as a member of a new family of eight-stranded beta-sheet proteins by its sequence and properties
Abstract
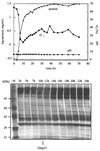
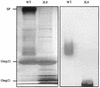
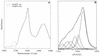
Omp21, a minor outer membrane protein of the soil bacterium Comamonas acidovorans, was purified from a spontaneous mutant lacking a surface layer and long-chain lipopolysaccharide. Omp21 synthesis is enhanced by oxygen depletion, and the protein has a variable electrophoretic mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis due to its heat-modifiable behavior. The structural gene omp21 encodes a precursor of 204 amino acids with a putative signal peptide of 21 amino acids. Mature Omp21 is a typical outer membrane protein with a high content of beta structure as determined by infrared spectroscopy. Sequence comparisons show that it belongs to a new outer membrane protein family, characterized by eight amphipathic beta strands, which includes virulence proteins, such as the neisserial opacity proteins, Salmonella typhimurium Rck, and Yersinia enterocolitica Ail, as well as the major outer membrane proteins OmpA from Escherichia coli and OprF from Pseudomonas aeruginosa.
Figures




References
-
- Applegate M, Juhn G, Liechty M, Moore M, Hozier J. Use of DNA purified in situ from cells embedded in agarose plugs for the molecular analysis of tk−/− mutants recovered in the L5178Y tk+/− 3.7.2C mutagen assay system. Mutat Res. 1990;245:55–59. - PubMed
-
- Baldermann, C., and H. Engelhardt. Unpublished data.
-
- Barondess J J, Beckwith J. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature. 1990;346:871–874. - PubMed
-
- Bhat K S, Gibbs C P, Barrera O, Morrison S G, Jahnig F, Stern A, Kupsch E M, Meyer T F, Swanson J. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol Microbiol. 1991;5:1889–1901. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

