Physical and genetic map of the obligate intracellular bacterium Coxiella burnetii
- PMID: 9683477
- PMCID: PMC107364
- DOI: 10.1128/JB.180.15.3816-3822.1998
Physical and genetic map of the obligate intracellular bacterium Coxiella burnetii
Abstract
Pulsed-field gel electrophoresis and PCR techniques have been used to construct a NotI macrorestriction map of the obligate intracellular bacterium Coxiella burnetii Nine Mile. The size of the chromosome has been determined to be 2,103 kb comprising 29 NotI restriction fragments. The average resolution is 72.5 kb, or about 3. 5% of the genome. Experimental data support the presence of a linear chromosome. Published genes were localized on the physical map by Southern hybridization. One gene, recognized as transposable element, was found to be present in at least nine sites evenly distributed over the whole chromosome. There is only one copy of a 16S rRNA gene. The putative oriC has been located on a 27.5-kb NotI fragment. Gene organization upstream the oriC is almost identical to that of Pseudomonas putida and Bacillus subtilis, whereas gene organization downstream the oriC seems to be unique among bacteria. The physical map will be helpful in investigations of the great heterogeneity in restriction fragment length polymorphism patterns of different isolates and the great variation in genome size. The genetic map will help to determine whether gene order in different isolates is conserved.
Figures


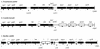
References
-
- Afseth G, Mallavia L P. Copy number of the 16S rRNA gene in Coxiella burnetii. Eur J Epidemiol. 1997;13:729–731. - PubMed
-
- Aitken I D, Bögel K, Cracera E, Edlinger E, Houwers D, Krauss H, Rady M, Rehacek J, Schiefer H G, Schmeer N, Tarasevich I V, Tringali G. Q fever in Europe: current aspects of aetiology, epidemiology, human infection, diagnosis and therapy (report of WHO Workshop on Q Fever) Infection. 1987;15:323–328. - PubMed
-
- Burger C. Ein rekombinantes Protein von Coxiella burnetii als Kandidat für eine Vakzine: Untersuchungen zum Vorkommen des omp-Gens und zur Biosynthese, Reinigung und Antigenität des rekombinanten OMP-Proteins. Vet. Med. thesis. Giessen, Germany: Justus-Liebig-Universität Giessen; 1996.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

