Purification and characterization of EDTA monooxygenase from the EDTA-degrading bacterium BNC1
- PMID: 9683478
- PMCID: PMC107365
- DOI: 10.1128/JB.180.15.3823-3827.1998
Purification and characterization of EDTA monooxygenase from the EDTA-degrading bacterium BNC1
Erratum in
- J Bacteriol 1998 Nov;180(21):5808
Abstract
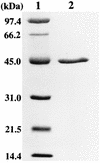
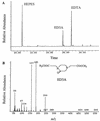
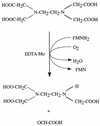
The synthetic chelating agent EDTA can mobilize radionuclides and heavy metals in the environment. Biodegradation of EDTA should reduce this mobilization. Although several bacteria have been reported to mineralize EDTA, little is known about the biochemistry of EDTA degradation. Understanding the biochemistry will facilitate the removal of EDTA from the environment. EDTA-degrading activities were detected in cell extracts of bacterium BNC1 when flavin mononucleotide (FMN), NADH, and O2 were present. The degradative enzyme system was separated into two different enzymes, EDTA monooxygenase and an FMN reductase. EDTA monooxygenase oxidized EDTA to glyoxylate and ethylenediaminetriacetate (ED3A), with the coconsumption of FMNH2 and O2. The FMN reductase provided EDTA monooxygenase with FMNH2 by reducing FMN with NADH. The FMN reductase was successfully substituted in the assay mixture by other FMN reductases. EDTA monooxygenase was purified to greater than 95% homogeneity and had a single polypeptide with a molecular weight of 45,000. The enzyme oxidized both EDTA complexed with various metal ions and uncomplexed EDTA. The optimal conditions for activity were pH 7.8 and 35 degreesC. Kms were 34.1 microM for uncomplexed EDTA and 8.5 microM for MgEDTA2-; this difference in Km indicates that the enzyme has greater affinity for MgEDTA2-. The enzyme also catalyzed the release of glyoxylate from nitrilotriacetate and diethylenetriaminepentaacetate. EDTA monooxygenase belongs to a small group of FMNH2-utilizing monooxygenases that attack carbon-nitrogen, carbon-sulfur, and carbon-carbon double bonds.
Figures
Similar articles
-
Cloning, sequencing, and characterization of a gene cluster involved in EDTA degradation from the bacterium BNC1.Appl Environ Microbiol. 2001 Feb;67(2):688-95. doi: 10.1128/AEM.67.2.688-695.2001. Appl Environ Microbiol. 2001. PMID: 11157232 Free PMC article.
-
Identification, purification, and characterization of iminodiacetate oxidase from the EDTA-degrading bacterium BNC1.Appl Environ Microbiol. 2001 Feb;67(2):696-701. doi: 10.1128/AEM.67.2.696-701.2001. Appl Environ Microbiol. 2001. PMID: 11157233 Free PMC article.
-
Identification and characterization of the two-enzyme system catalyzing oxidation of EDTA in the EDTA-degrading bacterial strain DSM 9103.J Bacteriol. 1997 Nov;179(22):6937-43. doi: 10.1128/jb.179.22.6937-6943.1997. J Bacteriol. 1997. PMID: 9371437 Free PMC article.
-
Structural and biochemical characterization of EDTA monooxygenase and its physical interaction with a partner flavin reductase.Mol Microbiol. 2016 Jun;100(6):989-1003. doi: 10.1111/mmi.13363. Epub 2016 Apr 13. Mol Microbiol. 2016. PMID: 26928990 Free PMC article.
-
Environmental fate and microbial degradation of aminopolycarboxylic acids.FEMS Microbiol Rev. 2001 Jan;25(1):69-106. doi: 10.1111/j.1574-6976.2001.tb00572.x. FEMS Microbiol Rev. 2001. PMID: 11152941 Review.
Cited by
-
Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new Flavin:NAD(P)H reductase subfamily.J Bacteriol. 2000 Feb;182(3):627-36. doi: 10.1128/JB.182.3.627-636.2000. J Bacteriol. 2000. PMID: 10633095 Free PMC article.
-
Identification of the rctA gene, which is required for repression of conjugative transfer of rhizobial symbiotic megaplasmids.J Bacteriol. 2005 Nov;187(21):7341-50. doi: 10.1128/JB.187.21.7341-7350.2005. J Bacteriol. 2005. PMID: 16237017 Free PMC article.
-
Cloning, sequencing, and characterization of a gene cluster involved in EDTA degradation from the bacterium BNC1.Appl Environ Microbiol. 2001 Feb;67(2):688-95. doi: 10.1128/AEM.67.2.688-695.2001. Appl Environ Microbiol. 2001. PMID: 11157232 Free PMC article.
-
A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism.J Biol Chem. 2010 Jul 16;285(29):22264-75. doi: 10.1074/jbc.M109.099028. Epub 2010 May 6. J Biol Chem. 2010. PMID: 20448045 Free PMC article.
-
The Role of Dioxygen in Microbial Bio-Oxygenation: Challenging Biochemistry, Illustrated by a Short History of a Long Misunderstood Enzyme.Microorganisms. 2024 Feb 15;12(2):389. doi: 10.3390/microorganisms12020389. Microorganisms. 2024. PMID: 38399793 Free PMC article. Review.
References
-
- Allison J D, Brown D S, Novo-Gradac K J. MinteqA2/ProdefA2, a geochemical assessment model for environmental systems: version 3.0 users manual. EPA/600/3-91/021. Athens, Ga: Environmental Protection Agency; 1991.
-
- Avers J A. Decontamination of nuclear reactors and equipment. New York, N.Y: The Ronald Ress Co.; 1970.
-
- Bolton H, Jr, Li S W, Workman D J, Girvin D C. Biodegradation of synthetic chelates in subsurface sediments from the Southeast coastal plain. J Environ Qual. 1993;22:125–132.
-
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Campbell J A. Flammable gas safety program. Analytical methods development: FY 1993 Progress Report. PNL-9062. Richland, Wash: Pacific Northwest National Laboratory; 1994.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

