Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR
- PMID: 9683482
- PMCID: PMC107369
- DOI: 10.1128/JB.180.15.3853-3863.1998
Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR
Abstract
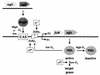
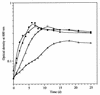
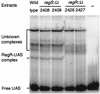
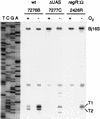
Many nitrogen fixation-associated genes in the soybean symbiont Bradyrhizobium japonicum are regulated by the transcriptional activator NifA, whose activity is inhibited by aerobiosis. NifA is encoded in the fixR-nifA operon, which is expressed at a low level under aerobic conditions and induced approximately fivefold under low-oxygen tension. This induction depends on a -24/-12-type promoter (fixRp1) that is recognized by the sigma54 RNA polymerase and activated by NifA. Low-level aerobic expression and part of the anaerobic expression originates from a second promoter (fixRp2) that overlaps with fixRp1 and depends on an upstream DNA region (UAS) located around position -68 (H. Barrios, H. M. Fischer, H. Hennecke, and E. Morett, J. Bacteriol. 177:1760-1765, 1995). A protein binding to the UAS was previously postulated to act as an activator. This protein has now been purified, and the corresponding gene (regR) has been cloned. On the basis of the predicted amino acid sequence, RegR belongs to the family of response regulators of two-component regulatory systems. We identified upstream of the regR gene an additional gene (regS) encoding a putative sensor kinase. A regR mutant was constructed in which neither a specific UAS-binding activity nor fixRp2-dependent transcript formation and fixR'-'lacZ expression was detected in aerobically grown cells. Anaerobic fixR'-'lacZ expression was also decreased in regR mutants to about 10% of the level observed in the wild type. Similarly, regR mutants showed only about 2% residual nitrogen fixation activity, but unlike nodules induced by nifA mutants, the morphology of those nodules was normal, displaying no signs of necrosis. While regR mutants grew only slightly slower in free-living, aerobic conditions, they displayed a strong growth defect under anaerobic conditions. The phenotypic properties of regS mutants differed only marginally, if at all, from those of the wild type, suggesting the existence of a compensating sensor activity in these strains. The newly identified RegR protein may be regarded as a master regulator in the NifA-dependent network controlling nif and fix gene expression in B. japonicum.
Figures
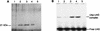




References
-
- Agron P G, Helinski D R. Symbiotic expression of Rhizobium meliloti nitrogen fixation genes is regulated by oxygen. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 275–287.
-
- Babst M, Hennecke H, Fischer H M. Two different mechanisms are involved in the heat shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. - PubMed
-
- Baker M E. Human placental 17β-hydroxysteroid dehydrogenase is homologous to NodG protein of Rhizobium meliloti. Mol Endocrinol. 1989;3:881–884. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

