Control by nutrients of growth and cell cycle progression in budding yeast, analyzed by double-tag flow cytometry
- PMID: 9683483
- PMCID: PMC107370
- DOI: 10.1128/JB.180.15.3864-3872.1998
Control by nutrients of growth and cell cycle progression in budding yeast, analyzed by double-tag flow cytometry
Abstract
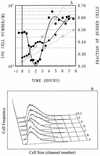
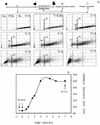
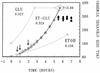
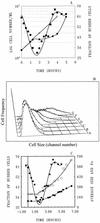
To gain insight on the interrelationships of the cellular environment, the properties of growth, and cell cycle progression, we analyzed the dynamic reactions of individual Saccharomyces cerevisiae cells to changes and manipulations of their surroundings. We used a new flow cytometric approach which allows, in asynchronous growing S. cerevisiae populations, tagging of both the cell age and the cell protein content of cells belonging to the different cell cycle set points. Since the cell protein content is a good estimation of the cell size, it is possible to follow the kinetics of the cell size increase during cell cycle progression. The analysis of the findings obtained indicates that both during a nutritional shift-up (from ethanol to glucose) and following the addition of cyclic AMP (cAMP), two important delays are induced. The preexisting cells that at the moment of the nutritional shift-up were cycling before the Start phase delay their entrance into S phase, while cells that were cycling after Start are delayed in their exit from the cycle. The combined effects of the two delays allow the cellular population that preexisted the shift-up to quickly adjust to the new growth condition. The effects of a nutritional shift-down were also determined.
Figures

References
-
- Alberghina L, Mariani L, Martegani E, Vanoni M. Analysis of protein distribution in budding yeast. Biotechnol Bioeng. 1983;25:1295–1310. - PubMed
-
- Alberghina L, Martegani E, Mariani L, Bortolan G. A bimolecular mechanism for the cell size control of the cell cycle. BioSystems. 1984;16:297–305. - PubMed
-
- Alberghina L, Porro D. Quantitative flow cytometry: analysis of protein distributions in budding yeast. A mini-review. Yeast. 1993;9:815–823. - PubMed
-
- Alberghina L, Ranzi B M, Porro D, Martegani E. Flow cytometry and cell size kinetics in continuous and fed-batch fermentations of budding yeast. Biotechnol Prog. 1991;7:299–304. - PubMed
-
- Ballou C E. Yeast cell wall and cell surface: metabolism and gene expression. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 335–360.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

