A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis
- PMID: 9683488
- PMCID: PMC107375
- DOI: 10.1128/JB.180.15.3907-3916.1998
A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis
Abstract
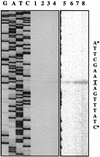
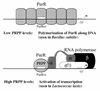
A purR::pGh9:ISS1 mutant of Lactococcus lactis was obtained following transposon mutagenesis of strain MG1363 and selection for purine auxotrophs. After determination of the nucleotide sequence and deduction of the purR reading frame, the PurR product was found to be highly similar to the purR-encoded repressor from Bacillus subtilis. The wild-type purR gene complemented the purine auxotrophy of a purR::ISS1 mutant, and it was shown that the purR::ISS1 mutation lowered the level of transcription from the purine-regulated L. lactis purD promoter. In a parallel study on the regulation of purC and purD expression in L. lactis (M. Kilstrup, S. G. Jessing, S. B. Wichmand-Jorgensen, M. Madsen, and D. Nilsson, J. Bacteriol. 180:3900-3906, 1998), we identified regions (PurBox sequences: AWWWCCGAACWWT) upstream of the promoters with a central G residue at exactly position -76 relative to the transcriptional start site. The PurBox sequences were found to be required for high-level promoter activity and purine regulation. We identified a PurBox sequence overlapping the -35 region of the L. lactis purR promoter and found, by studies of a purR-lacLM fusion plasmid, that purR is autoregulated. Because of the high degree of similarity of the PurR proteins from B. subtilis and L. lactis, we looked for PurBox sequences in the promoter regions of the PurR-regulated genes in B. subtilis and identified a perfectly matching PurBox sequence in the purA promoter region and slightly degenerate PurBox-like sequences in the promoter regions for the pur operon and the purR gene. Interestingly, the PurBox in the pur operon of B. subtilis is located almost identically, with respect to the promoter, to the PurBox sequences located in front of purC and purD in L. lactis. We present a hypothesis to explain how an ancestral PurR protein in B. subtilis could have evolved from an activator of the pur operon into a repressor which regulates transcription initiation from the same pur promoter by using the same PurR binding site and a similar response toward its effectors.
Figures


Similar articles
-
Activation control of pur gene expression in Lactococcus lactis: proposal for a consensus activator binding sequence based on deletion analysis and site-directed mutagenesis of purC and purD promoter regions.J Bacteriol. 1998 Aug;180(15):3900-6. doi: 10.1128/JB.180.15.3900-3906.1998. J Bacteriol. 1998. PMID: 9683487 Free PMC article.
-
The PurR regulon in Lactococcus lactis - transcriptional regulation of the purine nucleotide metabolism and translational machinery.Microbiology (Reading). 2012 Aug;158(Pt 8):2026-2038. doi: 10.1099/mic.0.059576-0. Epub 2012 Jun 7. Microbiology (Reading). 2012. PMID: 22679106
-
Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO.J Bacteriol. 2001 Nov;183(21):6175-83. doi: 10.1128/JB.183.21.6175-6183.2001. J Bacteriol. 2001. PMID: 11591660 Free PMC article.
-
Recombinant protein secretion by Bacillus subtilis and Lactococcus lactis: pathways, applications, and innovation potential.Essays Biochem. 2021 Jul 26;65(2):187-195. doi: 10.1042/EBC20200171. Essays Biochem. 2021. PMID: 33955475 Free PMC article. Review.
-
Organization and regulation of genes encoding biosynthetic enzymes in Bacillus subtilis.J Biol Chem. 1988 Feb 5;263(4):1595-8. J Biol Chem. 1988. PMID: 3123474 Review. No abstract available.
Cited by
-
Phosphoribosyl Diphosphate (PRPP): Biosynthesis, Enzymology, Utilization, and Metabolic Significance.Microbiol Mol Biol Rev. 2016 Dec 28;81(1):e00040-16. doi: 10.1128/MMBR.00040-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2016. PMID: 28031352 Free PMC article. Review.
-
PePPER: a webserver for prediction of prokaryote promoter elements and regulons.BMC Genomics. 2012 Jul 2;13:299. doi: 10.1186/1471-2164-13-299. BMC Genomics. 2012. PMID: 22747501 Free PMC article.
-
The hpx genetic system for hypoxanthine assimilation as a nitrogen source in Klebsiella pneumoniae: gene organization and transcriptional regulation.J Bacteriol. 2008 Dec;190(24):7892-903. doi: 10.1128/JB.01022-08. Epub 2008 Oct 10. J Bacteriol. 2008. PMID: 18849434 Free PMC article.
-
Lactococcus lactis Diversity in Undefined Mixed Dairy Starter Cultures as Revealed by Comparative Genome Analyses and Targeted Amplicon Sequencing of epsD.Appl Environ Microbiol. 2018 Jan 17;84(3):e02199-17. doi: 10.1128/AEM.02199-17. Print 2018 Feb 1. Appl Environ Microbiol. 2018. PMID: 29222100 Free PMC article.
-
Uracil salvage pathway in Lactobacillus plantarum: Transcription and genetic studies.J Bacteriol. 2006 Jul;188(13):4777-86. doi: 10.1128/JB.00195-06. J Bacteriol. 2006. PMID: 16788187 Free PMC article.
References
-
- Arnau J, Sørensen K I, Appel K F, Vogensen F K, Hammer K. Analysis of heat shock gene expression in Lactococcus lactis MG1363. Microbiology. 1996;142:1685–1691. - PubMed
-
- Arnvig K, Hove-Jensen B, Switzer R L. Purification and properties of phosphoribosyl-diphosphate synthetase from Bacillus subtilis. Eur J Biochem. 1990;192:195–200. - PubMed
-
- Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

