Identification of an intragenic ribosome binding site that affects expression of the uncB gene of the Escherichia coli proton-translocating ATPase (unc) operon
- PMID: 9683492
- PMCID: PMC107379
- DOI: 10.1128/JB.180.15.3940-3945.1998
Identification of an intragenic ribosome binding site that affects expression of the uncB gene of the Escherichia coli proton-translocating ATPase (unc) operon
Abstract
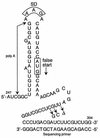
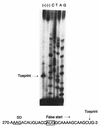
The uncB gene codes for the a subunit of the Fo proton channel sector of the Escherichia coli F1 Fo ATPase. Control of expression of uncB appears to be exerted at some step after translational initiation. Sequence analysis by the perceptron matrices (G. D. Stormo, T. D. Schneider, L. Gold, and A. Ehrenfeucht, Nucleic Acids Res. 10:2997-3011, 1982) identified a potential ribosome binding site within the uncB reading frame preceding a five-codon reading frame which is shifted one base relative to the uncB reading frame. Elimination of this binding site by mutagenesis resulted in a four- to fivefold increase in expression of an uncB'-'lacZ fusion gene containing most of uncB. Primer extension inhibition (toeprint) analysis to measure ribosome binding demonstrated that ribosomes could form an initiation complex at this alternative start site. Two fusions of lacZ to the alternative reading frame demonstrated that this site is recognized by ribosomes in vivo. The results suggest that expression of uncB is reduced by translational frameshifting and/or a translational false start at this site within the uncB reading frame.
Figures
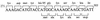



Similar articles
-
Effects of the uncI gene on expression of uncB, the gene coding for the a subunit of the F1F0 ATPase of Escherichia coli.FEBS Lett. 1995 Sep 4;371(2):127-31. doi: 10.1016/0014-5793(95)00867-9. FEBS Lett. 1995. PMID: 7672111
-
Degradation of Escherichia coli uncB mRNA by multiple endonucleolytic cleavages.J Bacteriol. 1995 Jul;177(14):3917-22. doi: 10.1128/jb.177.14.3917-3922.1995. J Bacteriol. 1995. PMID: 7608061 Free PMC article.
-
Use of lacZ fusions to measure in vivo expression of the first three genes of the Escherichia coli unc operon.J Bacteriol. 1989 Jun;171(6):3039-45. doi: 10.1128/jb.171.6.3039-3045.1989. J Bacteriol. 1989. PMID: 2524469 Free PMC article.
-
Inhibition of translation initiation on Escherichia coli gnd mRNA by formation of a long-range secondary structure involving the ribosome binding site and the internal complementary sequence.J Bacteriol. 1995 Nov;177(22):6560-7. doi: 10.1128/jb.177.22.6560-6567.1995. J Bacteriol. 1995. PMID: 7592434 Free PMC article.
-
Extremely Low Leakage Expression Systems Using Dual Transcriptional-Translational Control for Toxic Protein Production.Int J Mol Sci. 2020 Jan 21;21(3):705. doi: 10.3390/ijms21030705. Int J Mol Sci. 2020. PMID: 31973139 Free PMC article. Review.
Cited by
-
Time-delayed in vivo assembly of subunit a into preformed Escherichia coli FoF1 ATP synthase.J Bacteriol. 2013 Sep;195(18):4074-84. doi: 10.1128/JB.00468-13. Epub 2013 Jul 8. J Bacteriol. 2013. PMID: 23836871 Free PMC article.
-
Acquired gentamicin resistance by permeability impairment in Enterococcus faecalis.Antimicrob Agents Chemother. 2006 Nov;50(11):3615-21. doi: 10.1128/AAC.00390-06. Antimicrob Agents Chemother. 2006. PMID: 17065620 Free PMC article.
-
Twenty Years of Delila and Molecular Information Theory: The Altenberg-Austin Workshop in Theoretical Biology Biological Information, Beyond Metaphor: Causality, Explanation, and Unification Altenberg, Austria, 11-14 July 2002.Biol Theory. 2006;1(3):250-260. doi: 10.1162/biot.2006.1.3.250. Biol Theory. 2006. PMID: 18084638 Free PMC article.
-
Structure, Regulation, and Significance of Cyanobacterial and Chloroplast Adenosine Triphosphate Synthase in the Adaptability of Oxygenic Photosynthetic Organisms.Microorganisms. 2024 May 6;12(5):940. doi: 10.3390/microorganisms12050940. Microorganisms. 2024. PMID: 38792770 Free PMC article. Review.
-
In polycistronic Qbeta RNA, single-strandedness at one ribosome binding site directly affects translational initiations at a distal upstream cistron.Nucleic Acids Res. 2010 Nov;38(20):7199-210. doi: 10.1093/nar/gkq541. Epub 2010 Jun 25. Nucleic Acids Res. 2010. PMID: 20581118 Free PMC article.
References
-
- Foster D L, Fillingame R H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982;257:2009–2015. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

