Genetic analysis of dioxin dioxygenase of Sphingomonas sp. Strain RW1: catabolic genes dispersed on the genome
- PMID: 9683494
- PMCID: PMC107381
- DOI: 10.1128/JB.180.15.3954-3966.1998
Genetic analysis of dioxin dioxygenase of Sphingomonas sp. Strain RW1: catabolic genes dispersed on the genome
Abstract
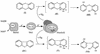
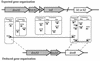
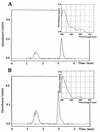
The dioxin dioxygenase of Sphingomonas sp. strain RW1 activates dibenzo-p-dioxin and dibenzofuran for further metabolism by introducing two atoms of oxygen at a pair of vicinal carbon atoms, one of which is involved in one of the bridges between the two aromatic rings, i.e., an angular dioxygenation. The dxnA1 and dxnA2 cistrons encoding this dioxygenase have been cloned and shown to be located just upstream of a hydrolase gene which specifies an enzyme involved in the subsequent step of the dibenzofuran biodegradative pathway. Genes encoding the electron supply system of the dioxygenase are not clustered with the dioxygenase gene but rather are located on two other distinct and separate genome segments. Moreover, whereas expression of dxnA1A2 is modulated according to the available carbon source, expression of the dbfB gene encoding the ring cleavage enzyme of the dibenzofuran pathway, which is located in the neighborhood of dxnA1A2 but oriented in the opposite direction, is constitutive. The scattering of genes for the component proteins of dioxin dioxygenase system around the genome of Sphingomonas sp. strain RW1, and the differential expression of dioxin pathway genes, is unusual and contrasts with the typical genetic organization of catabolic pathways where component cistrons tend to be clustered in multicistronic transcriptional units. The sequences of the alpha and beta subunits of the dioxin dioxygenase exhibit only weak similarity to other three component dioxygenases, but some motifs such as the Fe(II) binding site and the [2Fe-2S] cluster ligands are conserved. Dioxin dioxygenase activity in Escherichia coli cells containing the cloned dxnA1A2 gene was achieved only through coexpression of the cognate electron supply system from RW1. Under these conditions, exclusively angular dioxygenation of dibenzofuran and dibenzo-p-dioxin was obtained. The dioxin dioxygenase was not active in E. coli cells coexpressing a class IIB electron supply system. In the course of the isolation of the dxnA1 and dxnA2 cistrons, a number of other catabolic genes dispersed over different genome segments were identified, which may indicate greater catabolic potential than was previously suspected. This finding is consistent with the catabolic versatility of members of the genus Sphingomonas, which is becoming increasingly evident, and may indicate a less well evolved and regulated but more dynamic genetic organization in this organism than is the case for better-studied pathways in organisms such as Pseudomonas species.
Figures



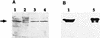

References
-
- Armengaud J, Timmis K N. Molecular characterization of Fdx1, a putidaredoxin-type [2Fe-2S] ferredoxin able to transfer electrons to the dioxin dioxygenase of Sphingomonas sp. RW1. Eur J Biochem. 1997;247:833–842. - PubMed
-
- Armengaud J, Timmis K N. The reductase RedA2 of the multicomponent dioxin dioxygenase system of Sphingomonas sp. RW1 is related to class-I cytochrome P450-type reductases. Eur J Biochem. 1998;253:437–444. - PubMed
-
- Asturias J A, Diaz E, Timmis K N. The evolutionary relationship of biphenyl dioxygenase from gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from gram-negative bacteria. Gene. 1995;156:11–18. - PubMed
-
- Batie C J, Ballou D P, Correll C C. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases. In: Müller F, editor. Chemistry and biochemistry of flavoenzymes. Boca Raton, Fla: CRC Press; 1992. pp. 543–566.
-
- Beil S, Happe B, Timmis K N, Pieper D H. Genetic and biochemical characterization of the broad spectrum chlorobenzene dioxygenase from Burkholderia sp. strain PS12: dechlorination of 1,2,4,5-tetrachlorobenzene. Eur J Biochem. 1997;247:190–199. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

