Transcription of the sodium/myo-inositol cotransporter gene is regulated by multiple tonicity-responsive enhancers spread over 50 kilobase pairs in the 5'-flanking region
- PMID: 9685419
- PMCID: PMC2365891
- DOI: 10.1074/jbc.273.32.20615
Transcription of the sodium/myo-inositol cotransporter gene is regulated by multiple tonicity-responsive enhancers spread over 50 kilobase pairs in the 5'-flanking region
Abstract
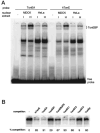
The sodium/myo-inositol cotransporter is a plasma membrane protein responsible for concentrative cellular accumulation of myo-inositol in a variety of tissues. When cells in kidney and brain are exposed to a hyperosmolar salt condition (hypertonicity) due to the operation of urinary concentration mechanism and pathological conditions, respectively, they survive the stress of hypertonicity by raising the cellular concentration of myo-inositol. Transcription of the sodium/myo-inositol cotransporter gene is markedly stimulated in response to hypertonicity, leading to an increase in the activity of the cotransporter, which in turn drives the osmoprotective accumulation of myo-inositol. To understand the molecular mechanisms by which hypertonicity stimulates transcription, we analyzed the 5'-flanking region of the cotransporter gene for cis-acting regulatory sequences. We identified five tonicity-responsive enhancers that are scattered over 50 kilobase pairs. All the enhancers are variations of the same type of enhancer interacting with the transcription factor named tonicity-responsive enhancer binding protein. In vivo methylation experiments demonstrated that exposure of cells to hypertonicity increases the binding of tonicity-responsive enhancer binding protein to the enhancer sites, indicating that all of these enhancers are involved in the transcriptional stimulation. We conclude that the sodium/myo-inositol cotransporter gene is regulated by a large region (approximately 50 kilobase pairs) upstream of the gene.
Figures



References
-
- Kwon HM, Yamauchi A, Uchida S, Preston AS, Garcia-Perez A, Burg MB, Handler JS. J Biol Chem. 1992;267:6297–6301. - PubMed
-
- Hager K, Hazama A, Kwon HM, Loo DDF, Handler JS, Wright EM. J Membr Biol. 1995;143:103–113. - PubMed
-
- Yamauchi A, Nakanishi T, Takamitsu Y, Sugita M, Imai E, Noguchi T, Fujiwara Y, Kamada T, Ueda N. J Am Soc Nephrol. 1994;5:62–67. - PubMed
-
- Yamauchi A, Uchida S, Preston AS, Kwon HM, Handler JS. Am J Physiol. 1993;264:F20–F23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

