Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics
- PMID: 9687389
- PMCID: PMC105715
- DOI: 10.1128/AAC.42.8.1952
Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics
Abstract
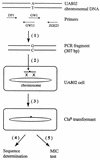
Clarithromycin resistance in Helicobacter pylori is mainly due to A-to-G mutations within the peptidyltransferase region of the 23S rRNA. In the present study, cross-resistance to macrolide, lincosamide, and streptogramin B (MLS) antibiotics (MLS phenotypes) has been investigated for several clinical isolates of H. pylori. Two major types of MLS resistance were identified and correlated with specific point mutations in the 23S rRNA gene. The A2142G mutation was linked with high-level cross-resistance to all MLS antibiotics (type I), and the A2143G mutation gave rise to an intermediate level of resistance to clarithromycin and clindamycin but no resistance to streptogramin B (type II). In addition, streptogramin A and streptogramin B were demonstrated to have a synergistic effect on both MLS-sensitive and MLS-resistant H. pylori strains. To further understand the mechanism of MLS resistance in H. pylori, we performed in vitro site-directed mutagenesis (substitution of G, C, or T for A at either position 2142 or 2143 of the 23S rRNA gene). The site-directed point mutations were introduced into a clarithromycin-susceptible strain, H. pylori UA802, by natural transformation followed by characterization of their effects on MLS resistance in an isogenic background. Strains with A-to-G and A-to-C mutations at the same position within the 23S rRNA gene had similar levels of clarithromycin resistance, and this level of resistance was higher than that for strains with the A-to-T mutation. Mutations at position 2142 conferred a higher level of clarithromycin resistance than mutations at position 2143. All mutations at position 2142 conferred cross-resistance to all MLS antibiotics, which corresponds to the type I MLS phenotype, whereas mutations at position 2143 were associated with a type II MLS phenotype with no resistance to streptogramin B. To explain that A-to-G transitions were predominantly observed in clarithromycin-resistant clinical isolates, we propose a possible mechanism by which A-to-G mutations are preferentially produced in H. pylori.
Figures


References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1991. pp. 8.5.7–8.5.9.
-
- Bryson H M, Spencer C M. Quinupristin-dalfopristin. Drugs. 1996;52:406–415. - PubMed
-
- Cayla, R., F. Zerbib, P. Talbi, F. Megraud, and H. Lamouliatte. 1995. Pre- and posttreatment clarithromycin resistance of Helicobacter pylori strains: a key factor of treatment failure. Gut 37(Suppl. 1):A55.
-
- Cocito, C., M. D. Giambattista, E. Nyssen, and P. Vannuffel. 1997. Inhibition of protein synthesis by streptogramins and related antibiotics. J. Antimicrob. Chemother. 39(Suppl. A):7–13. - PubMed
-
- Cundliffe E. Recognition sites for antibiotics within rRNA. In: Hill W E, Dalberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C: American Society for Microbiology; 1990. pp. 479–490.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

