Genetic diversity of nifH gene sequences in paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments
- PMID: 9687429
- PMCID: PMC106771
- DOI: 10.1128/AEM.64.8.2770-2779.1998
Genetic diversity of nifH gene sequences in paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments
Abstract
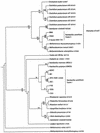
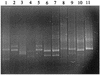
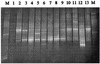
The diversity of dinitrogenase reductase gene (nifH) fragments in Paenibacillus azotofixans strains was investigated by using molecular methods. The partial nifH gene sequences of eight P. azotofixans strains, as well as one strain each of the close relatives Paenibacillus durum, Paenibacillus polymyxa, and Paenibacillus macerans, were amplified by PCR by using degenerate primers and were characterized by DNA sequencing. We found that there are two nifH sequence clusters, designated clusters I and II, in P. azotofixans. The data further indicated that there was sequence divergence among the nifH genes of P. azotofixans strains at the DNA level. However, the gene products were more conserved at the protein level. Phylogenetic analysis showed that all nifH cluster II sequences were similar to the alternative (anf) nitrogenase sequence. A nested PCR assay for the detection of nifH (cluster I) of P. azotofixans was developed by using the degenerate primers as outer primers and two specific primers, designed on the basis of the sequence information obtained, as inner primers. The specificity of the inner primers was tested with several diazotrophic bacteria, and PCR revealed that these primers are specific for the P. azotofixans nifH gene. A GC clamp was attached to one inner primer, and a denaturing gradient gel electrophoresis (DGGE) protocol was developed to study the genetic diversity of this region of nifH in P. azotofixans strains, as well as in soil and rhizosphere samples. The results revealed sequence heterogeneity among different nifH genes. Moreover, nifH is probably a multicopy gene in P. azotofixans. Both similarities and differences were detected in the P. azotofixans nifH DGGE profiles generated with soil and rhizosphere DNAs. The DGGE assay developed here is reproducible and provides a rapid way to assess the intraspecific genetic diversity of an important functional gene in pure cultures, as well as in environmental samples.
Figures



References
-
- Ash C, Farrow J A E, Priest F G, Collins M D. Molecular identification of rRNA group 3 bacilli using a PCR probe test. Proposal for the creation of a genus Paenibacillus. Antonie Leeuwenhoek. 1993;64:253–260. - PubMed
-
- Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. - PubMed
-
- Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 763–834.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

