Rac-1 regulates nuclear factor of activated T cells (NFAT) C1 nuclear translocation in response to Fcepsilon receptor type 1 stimulation of mast cells
- PMID: 9687530
- PMCID: PMC2212472
- DOI: 10.1084/jem.188.3.527
Rac-1 regulates nuclear factor of activated T cells (NFAT) C1 nuclear translocation in response to Fcepsilon receptor type 1 stimulation of mast cells
Abstract
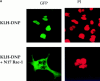
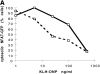
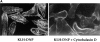
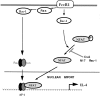
Transcription factors of the nuclear factor of activated T cells (NFAT) family play a key role in antigen receptor-mediated responses in lymphocytes by controlling induction of a wide variety of cytokine genes. The GTPases Ras and Rac-1 have essential functions in regulation of NFAT transcriptional activity in the mast cell system, where Fcepsilon receptor type 1 (FcepsilonR1) ligation results in induction of multiple NFAT target genes. This report examines the precise biochemical basis for the Rac-1 dependency of FcepsilonR1 activation of NFAT in mast cells. We are able to place Rac-1 in two positions in the signaling network that regulates the assembly and activation of NFAT transcriptional complexes in lymphocytes. First, we show that activity of Rac-1 is required for FcepsilonR1-mediated NFATC1 dephosphorylation and nuclear import. Regulation of NFAT localization by the FcepsilonR1 is a Rac-dependent but Ras-independent process. This novel signaling role for Rac-1 is distinct from its established regulation of the actin cytoskeleton. Our data also reveal a second GTPase signaling pathway regulating NFAT transcriptional activity, in which Rac-1 mediates a Ras signal. These data illustrate that the GTPase Rac-1 should now be considered as a component of the therapeutically important pathways controlling NFATC1 subcellular localization. They also reveal that GTPases may serve multiple functions in cellular responses to antigen receptor ligation.
Figures














References
-
- Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–234. - PubMed
-
- Jouvin MH, Numerof RP, Kinet JP. Signal transduction through the conserved motifs of the high affinity IgE receptor Fc epsilon RI. Semin Immunol. 1995;7:29–35. - PubMed
-
- Rao A, Luo C, Hogan P. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–748. - PubMed
-
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

