RNA-binding site in T7 RNA polymerase
- PMID: 9689042
- PMCID: PMC21300
- DOI: 10.1073/pnas.95.16.9111
RNA-binding site in T7 RNA polymerase
Abstract
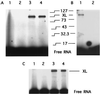
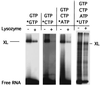
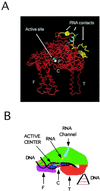
Recent models of RNA polymerase transcription complexes have invoked the idea that enzyme-nascent RNA contacts contribute to the stability of the complexes. Although much progress on this topic has been made with the multisubunit Escherichia coli RNA polymerase, there is a paucity of information regarding the structure of single-subunit phage RNA polymerase transcription complexes. Here, we photo-cross-linked the RNA in a T7 RNA polymerase transcription complex and mapped a major contact site between amino acid residues 144 and 168 and probably a minor contact between residues 1 and 93. These regions of the polymerase are proposed to interact with the emerging RNA during transcription because the 5' end of the RNA was cross-linked. The contacts are both ionic and nonionic (hydrophobic). The specific inhibitor of T7 transcription, T7 lysozyme, does not compete with T7 RNA polymerase for RNA cross-linking, implying that the RNA does not bind the lysozyme. However, lysozyme may act indirectly via a conformational change in the polymerase. In the current model, the DNA template lies in the polymerase cleft and the fingers subdomain may contact or maintain a template bubble, and a region in the N terminus forms a partly solvent-accessible binding channel for the emerging RNA.
Figures




References
-
- Uptain S M, Kane C, Chamberlin M. Annu Rev Biochem. 1997;66:117–172. - PubMed
-
- Burley S K, Roeder R G. Annu Rev Biochem. 1996;65:769–799. - PubMed
-
- Gamper H B, Hearst J E. Cell. 1982;29:81–90. - PubMed
-
- Yager T D, von Hippel P H. Biochemistry. 1991;30:1097–1118. - PubMed
-
- Chamberlin M J. Harvey Lect. 1993;88:1–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

