The Shake-and-Bake structure determination of triclinic lysozyme
- PMID: 9689072
- PMCID: PMC21330
- DOI: 10.1073/pnas.95.16.9284
The Shake-and-Bake structure determination of triclinic lysozyme
Abstract
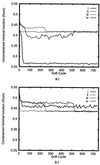
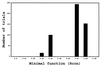
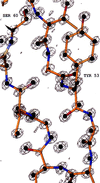
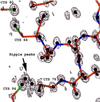
The crystal structure of triclinic lysozyme, comprised of 1,001 non-H protein atoms and approximately 200 bound water molecules, has been determined ab initio (using native data alone) by the "Shake-and-Bake" method by using the computer program SnB. This is the largest structure determined so far by the SnB program. Initial experiments, using default SnB parameters derived from studies of smaller molecules, were unsuccessful. In fact, such experiments produced electron density maps dominated by a single large peak. This problem was overcome by considering the choice of protocol used during the parameter-shift phase refinement. When each phase was subjected to a single shift of +/-157.5 degrees during each SnB cycle, an unusually high percentage of random trials (approximately 22%) yielded correct solutions within 750 cycles. This success rate is higher than that typically observed, even for much smaller structures.
Figures
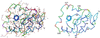
References
-
- Cochran W. Acta Crystallogr. 1955;8:473–478.
-
- Weeks C M, DeTitta G T, Miller R, Hauptman H A. Acta Crystallogr D. 1993;49:179–181. - PubMed
-
- Miller R, DeTitta G T, Jones R, Langs D A, Weeks C M, Hauptman H A. Science. 1993;259:1430–1433. - PubMed
-
- Weeks C M, DeTitta G T, Hauptman H A, Thuman P, Miller R. Acta Crystallogr A. 1994;50:210–220. - PubMed
-
- Miller R, Gallo S M, Khalak H G, Weeks C M. J Appl Crystallogr. 1994;27:613–621.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

