Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains
- PMID: 9693373
- PMCID: PMC25470
- DOI: 10.1091/mbc.9.8.2157
Mutations in Drosophila enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains
Abstract
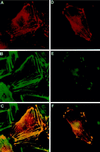
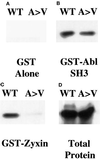
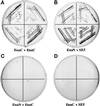
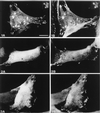
Drosophila Enabled (Ena) was initially identified as a dominant genetic suppressor of mutations in the Abelson tyrosine kinase and, more recently, as a member of the Ena/human vasodilator-stimulated phosphoprotein (VASP) family of proteins. We have used genetic, biochemical, and cell biological approaches to demonstrate the functional relationship between Ena and human VASP. In addition, we have defined the roles of Ena domains identified as essential for its activity in vivo. We have demonstrated that VASP rescues the embryonic lethality associated with loss of Ena function in Drosophila and have shown that Ena, like VASP, is associated with actin filaments and focal adhesions when expressed in cultured cells. To define sequences that are central to Ena function, we have characterized the molecular lesions present in two lethal ena mutant alleles that affected the Ena/VASP homology domain 1 (EVH1) and EVH2. A missense mutation that resulted in an amino acid substitution in the EVH1 domain eliminated in vitro binding of Ena to the cytoskeletal protein zyxin, a previously reported binding partner of VASP. A nonsense mutation that resulted in a C-terminally truncated Ena protein lacking the EVH2 domain failed to form multimeric complexes and exhibited reduced binding to zyxin and the Abelson Src homology 3 domain. Our analysis demonstrates that Ena and VASP are functionally homologous and defines the conserved EVH1 and EVH2 domains as central to the physiological activity of Ena.
Figures





References
-
- Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 1996;10:289–300. - PubMed
-
- Bennett RL, Hoffmann FM. Increased levels of the Drosophila Abelson tyrosine kinase in nerves and muscles: subcellular localization and mutant phenotypes imply a role in cell-cell interactions. Development. 1992;116:953–966. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

