Switch from translation to RNA replication in a positive-stranded RNA virus
- PMID: 9694795
- PMCID: PMC317040
- DOI: 10.1101/gad.12.15.2293
Switch from translation to RNA replication in a positive-stranded RNA virus
Abstract
In positive-stranded viruses, the genomic RNA serves as a template for both translation and RNA replication. Using poliovirus as a model, we examined the interaction between these two processes. We show that the RNA polymerase is unable to replicate RNA templates undergoing translation. We discovered that an RNA structure at the 5' end of the viral genome, next to the internal ribosomal entry site, carries signals that control both viral translation and RNA synthesis. The interaction of this RNA structure with the cellular factor PCBP up-regulates viral translation, while the binding of the viral protein 3CD represses translation and promotes negative-strand RNA synthesis. We propose that the interaction of 3CD with this RNA structure controls whether the genomic RNA is used for translation or RNA replication.
Figures
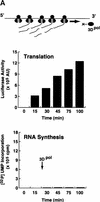

 ) into oocytes.
Luciferase activity was determined in oocyte extracts after 3 hr of
incubation at 22°C and expressed in AU. (D) Viral proteins
3Dpol, 3CD, and P3 copurified with the viral translation
inhibitory activity. Western blot analysis of fractions 1–14 eluted
from the HiTrap SP column is shown. Two microliters of each fraction
was resolved in a 10% SDS–polyacrylamide gel, transferred to
nitrocelluose membrane, and probed with specific anti-3CD antibodies.
The electrophoretic mobility of P3, 3CD, and 3D is indicated at
left.
) into oocytes.
Luciferase activity was determined in oocyte extracts after 3 hr of
incubation at 22°C and expressed in AU. (D) Viral proteins
3Dpol, 3CD, and P3 copurified with the viral translation
inhibitory activity. Western blot analysis of fractions 1–14 eluted
from the HiTrap SP column is shown. Two microliters of each fraction
was resolved in a 10% SDS–polyacrylamide gel, transferred to
nitrocelluose membrane, and probed with specific anti-3CD antibodies.
The electrophoretic mobility of P3, 3CD, and 3D is indicated at
left.
 ) into oocytes.
Luciferase activity was determined in oocyte extracts after 3 hr of
incubation at 22°C and expressed in AU. (D) Viral proteins
3Dpol, 3CD, and P3 copurified with the viral translation
inhibitory activity. Western blot analysis of fractions 1–14 eluted
from the HiTrap SP column is shown. Two microliters of each fraction
was resolved in a 10% SDS–polyacrylamide gel, transferred to
nitrocelluose membrane, and probed with specific anti-3CD antibodies.
The electrophoretic mobility of P3, 3CD, and 3D is indicated at
left.
) into oocytes.
Luciferase activity was determined in oocyte extracts after 3 hr of
incubation at 22°C and expressed in AU. (D) Viral proteins
3Dpol, 3CD, and P3 copurified with the viral translation
inhibitory activity. Western blot analysis of fractions 1–14 eluted
from the HiTrap SP column is shown. Two microliters of each fraction
was resolved in a 10% SDS–polyacrylamide gel, transferred to
nitrocelluose membrane, and probed with specific anti-3CD antibodies.
The electrophoretic mobility of P3, 3CD, and 3D is indicated at
left.
 ) into oocytes.
Luciferase activity was determined in oocyte extracts after 3 hr of
incubation at 22°C and expressed in AU. (D) Viral proteins
3Dpol, 3CD, and P3 copurified with the viral translation
inhibitory activity. Western blot analysis of fractions 1–14 eluted
from the HiTrap SP column is shown. Two microliters of each fraction
was resolved in a 10% SDS–polyacrylamide gel, transferred to
nitrocelluose membrane, and probed with specific anti-3CD antibodies.
The electrophoretic mobility of P3, 3CD, and 3D is indicated at
left.
) into oocytes.
Luciferase activity was determined in oocyte extracts after 3 hr of
incubation at 22°C and expressed in AU. (D) Viral proteins
3Dpol, 3CD, and P3 copurified with the viral translation
inhibitory activity. Western blot analysis of fractions 1–14 eluted
from the HiTrap SP column is shown. Two microliters of each fraction
was resolved in a 10% SDS–polyacrylamide gel, transferred to
nitrocelluose membrane, and probed with specific anti-3CD antibodies.
The electrophoretic mobility of P3, 3CD, and 3D is indicated at
left.
 ) into oocytes.
Luciferase activity was determined in oocyte extracts after 3 hr of
incubation at 22°C and expressed in AU. (D) Viral proteins
3Dpol, 3CD, and P3 copurified with the viral translation
inhibitory activity. Western blot analysis of fractions 1–14 eluted
from the HiTrap SP column is shown. Two microliters of each fraction
was resolved in a 10% SDS–polyacrylamide gel, transferred to
nitrocelluose membrane, and probed with specific anti-3CD antibodies.
The electrophoretic mobility of P3, 3CD, and 3D is indicated at
left.
) into oocytes.
Luciferase activity was determined in oocyte extracts after 3 hr of
incubation at 22°C and expressed in AU. (D) Viral proteins
3Dpol, 3CD, and P3 copurified with the viral translation
inhibitory activity. Western blot analysis of fractions 1–14 eluted
from the HiTrap SP column is shown. Two microliters of each fraction
was resolved in a 10% SDS–polyacrylamide gel, transferred to
nitrocelluose membrane, and probed with specific anti-3CD antibodies.
The electrophoretic mobility of P3, 3CD, and 3D is indicated at
left.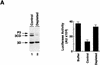
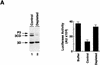
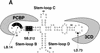
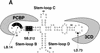
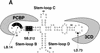
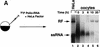
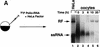
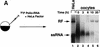
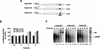
Similar articles
-
Interactions of viral protein 3CD and poly(rC) binding protein with the 5' untranslated region of the poliovirus genome.J Virol. 2000 Mar;74(5):2219-26. doi: 10.1128/jvi.74.5.2219-2226.2000. J Virol. 2000. PMID: 10666252 Free PMC article.
-
Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein.J Virol. 2007 Sep;81(17):8919-32. doi: 10.1128/JVI.01013-07. Epub 2007 Jun 20. J Virol. 2007. PMID: 17581994 Free PMC article.
-
Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro).J Virol. 2002 Mar;76(5):2529-42. doi: 10.1128/jvi.76.5.2529-2542.2002. J Virol. 2002. PMID: 11836431 Free PMC article.
-
Mechanistic intersections between picornavirus translation and RNA replication.Adv Virus Res. 2011;80:1-24. doi: 10.1016/B978-0-12-385987-7.00001-4. Adv Virus Res. 2011. PMID: 21762819 Review.
-
Molecular biology and cell-free synthesis of poliovirus.Biologicals. 1993 Dec;21(4):349-56. doi: 10.1006/biol.1993.1095. Biologicals. 1993. PMID: 8024750 Review.
Cited by
-
p33-Independent activation of a truncated p92 RNA-dependent RNA polymerase of Tomato bushy stunt virus in yeast cell-free extract.J Virol. 2012 Nov;86(22):12025-38. doi: 10.1128/JVI.01303-12. Epub 2012 Aug 29. J Virol. 2012. PMID: 22933278 Free PMC article.
-
Structure of the 5' nontranslated region of the coxsackievirus b3 genome: Chemical modification and comparative sequence analysis.J Virol. 2007 Jan;81(2):650-68. doi: 10.1128/JVI.01327-06. Epub 2006 Nov 1. J Virol. 2007. PMID: 17079314 Free PMC article.
-
Hepatitis A virus proteinase 3C binding to viral RNA: correlation with substrate binding and enzyme dimerization.Biochem J. 2005 Jan 15;385(Pt 2):363-70. doi: 10.1042/BJ20041153. Biochem J. 2005. PMID: 15361063 Free PMC article.
-
Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor.J Virol. 2005 Jun;79(11):7207-16. doi: 10.1128/JVI.79.11.7207-7216.2005. J Virol. 2005. PMID: 15890959 Free PMC article.
-
Cell-free synthesis of encephalomyocarditis virus.J Virol. 2003 Jun;77(11):6551-5. doi: 10.1128/jvi.77.11.6551-6555.2003. J Virol. 2003. PMID: 12743313 Free PMC article.
References
-
- Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990a;63:369–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
