A molecular switch controlling competence and motility: competence regulatory factors ComS, MecA, and ComK control sigmaD-dependent gene expression in Bacillus subtilis
- PMID: 9696775
- PMCID: PMC107423
- DOI: 10.1128/JB.180.16.4243-4251.1998
A molecular switch controlling competence and motility: competence regulatory factors ComS, MecA, and ComK control sigmaD-dependent gene expression in Bacillus subtilis
Abstract
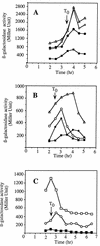
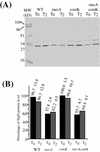
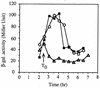
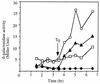
Bacillus subtilis, like many bacteria, will choose among several response pathways when encountering a stressful environment. Among the processes activated under growth-restricting conditions are sporulation, establishment of motility, and competence development. Recent reports implicate ComK and MecA-ClpC as part of a system that regulates both motility and competence development. MecA, while negatively controlling competence by inhibiting ComK, stimulates sigmaD-dependent transcription of genes that function in motility and autolysin production. Both ComK-dependent and -independent pathways have been proposed for MecA's role in the regulation of motility. Mutations in mecA reduce the transcription of hag. encoding flagellin, and are partially suppressed by comK in both medium promoting motility and medium promoting competence. Reduced sigmaD levels are observed in mecA mutants grown in competence medium, but no change in sigmaD concentration is detected in a comK mutant. The comF operon, transcription of which requires ComK, is located immediately upstream of the operon that contains the flgM gene, encoding the sigmaD-specific antisigma factor. An insertion mutation that disrupts the putative comF-flgM transcription unit confers a phenotype identical to that of the comK mutant with respect to hag-lacZ expression. Expression of a flgM-lacZ operon fusion is reduced in both sigD and comK mutant cells but is abolished in the sigD comK double mutant. Reverse transcription-PCR examination of the comF-flgM transcript indicates that readthrough from comF into the flgM operon is dependent on ComK. ComK negatively controls the transcription of hag by stimulating the transcription of comF-flgM, thereby increasing the production of the FlgM antisigma factor that inhibits sigmaD activity. There likely exists another comK-independent mechanism of hag transcription that requires mecA and possibly affects the sigmaD concentration in cells undergoing competence development.
Figures




Similar articles
-
Mutational analysis of ComS: evidence for the interaction of ComS and MecA in the regulation of competence development in Bacillus subtilis.Mol Microbiol. 1999 May;32(4):799-812. doi: 10.1046/j.1365-2958.1999.01399.x. Mol Microbiol. 1999. PMID: 10361283
-
Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis.J Bacteriol. 1998 Jul;180(14):3548-55. doi: 10.1128/JB.180.14.3548-3555.1998. J Bacteriol. 1998. PMID: 9657996 Free PMC article.
-
Effects of mecA and mecB (clpC) mutations on expression of sigD, which encodes an alternative sigma factor, and autolysin operons and on flagellin synthesis in Bacillus subtilis.J Bacteriol. 1996 Aug;178(16):4861-9. doi: 10.1128/jb.178.16.4861-4869.1996. J Bacteriol. 1996. PMID: 8759849 Free PMC article.
-
A regulatory switch involving a Clp ATPase.Bioessays. 1997 Jun;19(6):455-8. doi: 10.1002/bies.950190604. Bioessays. 1997. PMID: 9204762 Review.
-
Controlling competence in Bacillus subtilis: shared use of regulators.Microbiology (Reading). 2003 Jan;149(Pt 1):9-17. doi: 10.1099/mic.0.26003-0. Microbiology (Reading). 2003. PMID: 12576575 Review.
Cited by
-
The Surfactin-Like Lipopeptides From Bacillus spp.: Natural Biodiversity and Synthetic Biology for a Broader Application Range.Front Bioeng Biotechnol. 2021 Mar 2;9:623701. doi: 10.3389/fbioe.2021.623701. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 33738277 Free PMC article.
-
Transient growth requirement in Bacillus subtilis following the cessation of exponential growth.Appl Environ Microbiol. 2000 Mar;66(3):1220-2. doi: 10.1128/AEM.66.3.1220-1222.2000. Appl Environ Microbiol. 2000. PMID: 10698797 Free PMC article.
-
The induction of natural competence adapts staphylococcal metabolism to infection.Nat Commun. 2022 Mar 21;13(1):1525. doi: 10.1038/s41467-022-29206-7. Nat Commun. 2022. PMID: 35314690 Free PMC article.
-
Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach.Nucleic Acids Res. 2002 Dec 15;30(24):5517-28. doi: 10.1093/nar/gkf698. Nucleic Acids Res. 2002. PMID: 12490720 Free PMC article.
-
A unique open reading frame within the comX gene of Streptococcus mutans regulates genetic competence and oxidative stress tolerance.Mol Microbiol. 2015 May;96(3):463-82. doi: 10.1111/mmi.12948. Epub 2015 Mar 4. Mol Microbiol. 2015. PMID: 25620525 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D O, Smith J A, Seidman J G, Struhl K. PCR amplification of RNA under optimal conditions. In: Ausubel F M, et al., editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1998. pp. 15.4.1–15.4.2.
-
- Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 1993;7:139–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

