Recombination in the 5' leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization region
- PMID: 9696788
- PMCID: PMC109916
- DOI: 10.1128/JVI.72.9.6967-6978.1998
Recombination in the 5' leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization region
Abstract
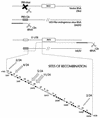
Retroviral recombination occurs frequently during reverse transcription of the dimeric RNA genome. By a forced recombination approach based on the transduction of Akv murine leukemia virus vectors harboring a primer binding site knockout mutation and the entire 5' untranslated region, we studied recombination between two closely related naturally occurring retroviral sequences. On the basis of 24 independent template switching events within a 481-nucleotide target sequence containing multiple sequence identity windows, we found that shifting from vector RNA to an endogenous retroviral RNA template during minus-strand DNA synthesis occurred within defined areas of the genome and did not lead to misincorporations at the crossover site. The nonrandom distribution of recombination sites did not reflect a bias for specific sites due to selection at the level of marker gene expression. We address whether template switching is affected by the length of sequence identity, by palindromic sequences, and/or by putative stem-loop structures. Sixteen of 24 sites of recombination colocalized with the kissing-loop dimerization region, and we propose that RNA-RNA interactions between palindromic sequences facilitate template switching. We discuss the putative role of the dimerization domain in the overall structure of the reverse-transcribed RNA dimer and note that related mechanisms of template switching may be found in remote RNA viruses.
Figures



Similar articles
-
Mutations of the kissing-loop dimerization sequence influence the site specificity of murine leukemia virus recombination in vivo.J Virol. 2000 Jan;74(2):600-10. doi: 10.1128/jvi.74.2.600-610.2000. J Virol. 2000. PMID: 10623721 Free PMC article.
-
Complementarity-directed RNA dimer-linkage promotes retroviral recombination in vivo.Nucleic Acids Res. 2004 Jan 9;32(1):102-14. doi: 10.1093/nar/gkh159. Print 2004. Nucleic Acids Res. 2004. PMID: 14715920 Free PMC article.
-
A preferred region for recombinational patch repair in the 5' untranslated region of primer binding site-impaired murine leukemia virus vectors.J Virol. 1996 Mar;70(3):1439-47. doi: 10.1128/JVI.70.3.1439-1447.1996. J Virol. 1996. PMID: 8627661 Free PMC article.
-
Dimerization of retroviral genomic RNAs: structural and functional implications.Biochimie. 1996;78(7):639-53. doi: 10.1016/s0300-9084(96)80010-1. Biochimie. 1996. PMID: 8955907 Review.
-
Recombination in Positive-Strand RNA Viruses.Front Microbiol. 2022 May 18;13:870759. doi: 10.3389/fmicb.2022.870759. eCollection 2022. Front Microbiol. 2022. PMID: 35663855 Free PMC article. Review.
Cited by
-
Duplication of the primary encapsidation and dimer linkage region of human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions.J Virol. 2001 Mar;75(6):2557-65. doi: 10.1128/JVI.75.6.2557-2565.2001. J Virol. 2001. PMID: 11222678 Free PMC article.
-
The kissing-loop motif is a preferred site of 5' leader recombination during replication of SL3-3 murine leukemia viruses in mice.J Virol. 1999 Nov;73(11):9614-8. doi: 10.1128/JVI.73.11.9614-9618.1999. J Virol. 1999. PMID: 10516072 Free PMC article.
-
Effect of the murine leukemia virus extended packaging signal on the rates and locations of retroviral recombination.J Virol. 2000 Aug;74(15):6953-63. doi: 10.1128/jvi.74.15.6953-6963.2000. J Virol. 2000. PMID: 10888634 Free PMC article.
-
Effect of distance between homologous sequences and 3' homology on the frequency of retroviral reverse transcriptase template switching.J Virol. 1999 Oct;73(10):7923-32. doi: 10.1128/JVI.73.10.7923-7932.1999. J Virol. 1999. PMID: 10482539 Free PMC article.
-
Mutations of the kissing-loop dimerization sequence influence the site specificity of murine leukemia virus recombination in vivo.J Virol. 2000 Jan;74(2):600-10. doi: 10.1128/jvi.74.2.600-610.2000. J Virol. 2000. PMID: 10623721 Free PMC article.
References
-
- Alford R L, Honda S, Lawrence C B, Belmont J W. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology. 1991;183:611–619. - PubMed
-
- Auerswald E A, Ludwig G, Schaller H. Structural analysis of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45:107–113. - PubMed
-
- Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

