Characterization of a baculovirus-encoded RNA 5'-triphosphatase
- PMID: 9696798
- PMCID: PMC109926
- DOI: 10.1128/JVI.72.9.7057-7063.1998
Characterization of a baculovirus-encoded RNA 5'-triphosphatase
Abstract
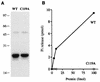
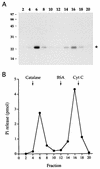
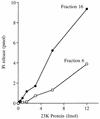
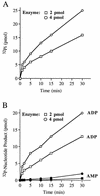
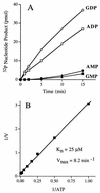
Autographa californica nuclear polyhedrosis virus (AcNPV) encodes a 168-amino-acid polypeptide that contains the signature motif of the superfamily of protein phosphatases that act via a covalent cysteinyl phosphate intermediate. The sequence of the AcNPV phosphatase is similar to that of the RNA triphosphatase domain of the metazoan cellular mRNA capping enzyme. Here, we show that the purified recombinant AcNPV protein is an RNA 5'-triphosphatase that hydrolyzes the gamma-phosphate of triphosphate-terminated poly(A); it also hydrolyzes ATP to ADP and GTP to GDP. The phosphatase sediments as two discrete components in a glycerol gradient: a 9.5S oligomer and 2.5S putative monomer. The 2.5S form of the enzyme releases 32Pi from 1 microM gamma-32P-labeled triphosphate-terminated poly(A) with a turnover number of 52 min-1 and converts ATP to ADP with Vmax of 8 min-1 and Km of 25 microM ATP. The 9.5S oligomeric form of the enzyme displays an initial pre-steady-state burst of ADP and Pi formation, which is proportional to and stoichiometric with the enzyme, followed by a slower steady-state rate of product formation (approximately 1/10 of the steady-state rate of the 2.5S enzyme). We surmise that the oligomeric enzyme is subject to a rate-limiting step other than reaction chemistry and that this step is either distinct from or slower than the rate-limiting step for the 2.5S enzyme. Replacing the presumptive active site nucleophile Cys-119 by alanine abrogates RNA triphosphatase and ATPase activity. Our findings raise the possibility that baculoviruses encode enzymes that cap the 5' ends of viral transcripts synthesized at late times postinfection by a virus-encoded RNA polymerase.
Figures


Similar articles
-
RNA 5'-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein.J Virol. 1998 Dec;72(12):10020-8. doi: 10.1128/JVI.72.12.10020-10028.1998. J Virol. 1998. PMID: 9811740 Free PMC article.
-
The LEF-4 subunit of baculovirus RNA polymerase has RNA 5'-triphosphatase and ATPase activities.J Virol. 1998 Dec;72(12):10011-9. doi: 10.1128/JVI.72.12.10011-10019.1998. J Virol. 1998. PMID: 9811739 Free PMC article.
-
Mutational analysis of the RNA triphosphatase component of vaccinia virus mRNA capping enzyme.J Virol. 1996 Sep;70(9):6162-8. doi: 10.1128/JVI.70.9.6162-6168.1996. J Virol. 1996. PMID: 8709242 Free PMC article.
-
Characterization of Schizosaccharomyces pombe RNA triphosphatase.Nucleic Acids Res. 2001 Jan 15;29(2):387-96. doi: 10.1093/nar/29.2.387. Nucleic Acids Res. 2001. PMID: 11139608 Free PMC article.
-
Yeast and viral RNA 5' triphosphatases comprise a new nucleoside triphosphatase family.J Biol Chem. 1998 Dec 18;273(51):34151-6. doi: 10.1074/jbc.273.51.34151. J Biol Chem. 1998. PMID: 9852075
Cited by
-
Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism.Proc Natl Acad Sci U S A. 1998 Oct 13;95(21):12226-31. doi: 10.1073/pnas.95.21.12226. Proc Natl Acad Sci U S A. 1998. PMID: 9770468 Free PMC article.
-
Magnesium-binding studies reveal fundamental differences between closely related RNA triphosphatases.Nucleic Acids Res. 2008 Feb;36(2):451-61. doi: 10.1093/nar/gkm1067. Epub 2007 Nov 26. Nucleic Acids Res. 2008. PMID: 18039706 Free PMC article.
-
RNA 5'-triphosphatase activity of the hepatitis E virus helicase domain.J Virol. 2010 Sep;84(18):9637-41. doi: 10.1128/JVI.00492-10. Epub 2010 Jun 30. J Virol. 2010. PMID: 20592074 Free PMC article.
-
Structure of AcMNPV nucleocapsid reveals DNA portal organization and packaging apparatus of circular dsDNA baculovirus.Nat Commun. 2025 May 24;16(1):4844. doi: 10.1038/s41467-025-60152-2. Nat Commun. 2025. PMID: 40413174 Free PMC article.
-
Roles of LEF-4 and PTP/BVP RNA triphosphatases in processing of baculovirus late mRNAs.J Virol. 2008 Jun;82(11):5573-83. doi: 10.1128/JVI.00058-08. Epub 2008 Apr 2. J Virol. 2008. PMID: 18385232 Free PMC article.
References
-
- Beniya H, Funk C J, Rohrmann G F, Weaver R F. Purification of a virus-induced RNA polymerase from Autographa californica nuclear polyhedrosis virus-infected Spodoptera frugiperda cells that accurately initiates late and very late transcription in vitro. Virology. 1996;216:12–19. - PubMed
-
- Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structures. Virology. 1997;236:1–7. - PubMed
-
- Carstens E B, Chan H, Yu H, Williams G V, Casselman R. Genetic analyses of temperature-sensitive mutations in baculovirus late expression genes. Virology. 1994;204:323–337. - PubMed
-
- Denu J M, Stuckey J A, Saper M A, Dixon J E. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

