The polyserine tract of herpes simplex virus ICP4 is required for normal viral gene expression and growth in murine trigeminal ganglia
- PMID: 9696805
- PMCID: PMC109933
- DOI: 10.1128/JVI.72.9.7115-7124.1998
The polyserine tract of herpes simplex virus ICP4 is required for normal viral gene expression and growth in murine trigeminal ganglia
Abstract
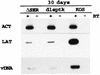
ICP4 of herpes simplex virus (HSV) is essential for productive infection due to its central role in the regulation of HSV transcription. This study identified a region of ICP4 that is not required for viral growth in culture or at the periphery of experimentally inoculated mice but is critical for productive growth in the trigeminal ganglia. This region of ICP4 encompasses amino acids 184 to 198 and contains 13 nearly contiguous serine residues that are highly conserved among the alphaherpesviruses. A mutant in which this region is deleted (DeltaSER) was able to grow on the corneas of mice and be transported back to the trigeminal ganglia. DeltaSER did not grow in the trigeminal ganglia but did express low levels of several immediate-early (ICP4 and ICP27) and early (thymidine kinase [tk] and UL42) genes. It expressed very low levels of the late gC gene and did not appear to replicate DNA. This pattern of gene expression was similar to that observed for a tk mutant, dlsptk. Both DeltaSER and dlsptk expressed higher levels of the latency-associated transcript (LAT) per genome earlier in infected ganglia than did the wild-type virus, KOS. However, infected ganglia from all three viruses accumulated the same level of LAT per genome at 30 days postinfection (during latency). The data suggest that the polyserine tract of ICP4 provides an activity that is required for lytic infection in ganglia to progress to viral DNA synthesis and full lytic gene expression. In the absence of this activity, higher levels of LAT per genome accumulate earlier in infection than with wild-type virus.
Figures







References
-
- Anderson A S, Francesconi A, Morgan R W. Complete nucleotide sequence of the Marek’s disease virus ICP4 gene. Virology. 1992;189:657–667. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

