Rotavirus RNA replication requires a single-stranded 3' end for efficient minus-strand synthesis
- PMID: 9696835
- PMCID: PMC109963
- DOI: 10.1128/JVI.72.9.7387-7396.1998
Rotavirus RNA replication requires a single-stranded 3' end for efficient minus-strand synthesis
Abstract
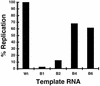
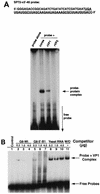
The segmented double-stranded (ds) RNA genome of the rotaviruses is replicated asymmetrically, with viral mRNA serving as the template for the synthesis of minus-strand RNA. Previous studies with cell-free replication systems have shown that the highly conserved termini of rotavirus gene 8 and 9 mRNAs contain cis-acting signals that promote the synthesis of dsRNA. Based on the location of the cis-acting signals and computer modeling of their secondary structure, the ends of the gene 8 or 9 mRNAs are proposed to interact in cis to form a modified panhandle structure that promotes the synthesis of dsRNA. In this structure, the last 11 to 12 nucleotides of the RNA, including the cis-acting signal that is essential for RNA replication, extend as a single-stranded tail from the panhandled region, and the 5' untranslated region folds to form a stem-loop motif. To understand the importance of the predicted secondary structure in minus-strand synthesis, mutations were introduced into viral RNAs which affected the 3' tail and the 5' stem-loop. Analysis of the RNAs with a cell-free replication system showed that, in contrast to mutations which altered the structure of the 5' stem-loop, mutations which caused complete or near-complete complementarity between the 5' end and the 3' tail significantly inhibited (>/=10-fold) minus-strand synthesis. Likewise, incubation of wild-type RNAs with oligonucleotides which were complementary to the 3' tail inhibited replication. Despite their replication-defective phenotype, mutant RNAs with complementary 5' and 3' termini were shown to competitively interfere with the replication of wild-type mRNA and to bind the viral RNA polymerase VP1 as efficiently as wild-type RNA. These results indicate that the single-strand nature of the 3' end of rotavirus mRNA is essential for efficient dsRNA synthesis and that the specific binding of the RNA polymerase to the mRNA template is required but not sufficient for the synthesis of minus-strand RNA.
Figures




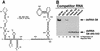

References
-
- Beekwilder M J, Nieuwenhuizen R, van Duin J. Secondary structure model for the last two domains of single-stranded RNA phage Q beta. J Mol Biol. 1995;247:903. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

