Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasispecies
- PMID: 9696836
- PMCID: PMC109966
- DOI: 10.1128/JVI.72.9.7397-7406.1998
Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasispecies
Abstract
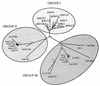
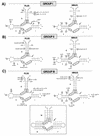
The peach latent mosaic viroid (PLMVd) is used to study the interactions between a viroid containing hammerhead ribozymes and its natural host, peach. To gain insight into the molecular basis of the phenotypic effects observed upon viroid infection, sequence variants from three PLMVd isolates that differ in symptom expression on the peach indicator GF-305 have been characterized. Analysis of the primary structures of a total of 29 different sequence variants derived from a severe and two latent isolates has revealed a large number of polymorphic positions in the viroid molecule. The variability pattern indicates that preservation of the stability of both hammerhead structures and conservation of a branched secondary structure of the viroid molecule may be factors limiting sequence heterogeneity in PLMVd. Moreover, compensatory mutations in two hairpin loops of the proposed secondary structure, suggesting that a pseudoknot-like interaction may exist between them, have also been observed. Phylogenetic analysis has allowed the allocation of PLMVd molecules into three major groups. This clustering does not strictly correlate with the source isolate from which the variants were obtained, providing insights into the complex mixture of molecules which make up each isolate. Bioassays of individual PLMVd sequence variants on GF-305 peach seedlings have shown that the biological properties of the PLMVd isolates may be correlated with both the complexity of their viroid populations and the presence of specific sequence variants.
Figures




References
-
- Ambrós S, Desvignes J C, Llácer G, Flores R. Peach latent mosaic and pear blister canker viroids: detection by molecular hybridization and relationships with specific maladies affecting peach and pear trees. Acta Hortic. 1995;386:515–521.
-
- Branch A D, Robertson H D. A replication cycle for viroids and other small infectious RNAs. Science. 1984;223:450–455. - PubMed
-
- Bruening G. Compilation of self-cleaving sequences from plant virus satellite RNAs and other sources. Methods Enzymol. 1989;180:546–558. - PubMed
-
- Côte F, Perrault J P. Peach latent mosaic viroid is locked by a 2′,5′-phosphodiester bond produced by in vitro self-ligation. J Mol Biol. 1997;273:533–543. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

